Crystal structure of the thioesterification conformation of Bacillus subtilis o-succinylbenzoyl-CoA synthetase reveals a distinct substrate-binding mode
- PMID: 28559280
- PMCID: PMC5519377
- DOI: 10.1074/jbc.M117.790410
Crystal structure of the thioesterification conformation of Bacillus subtilis o-succinylbenzoyl-CoA synthetase reveals a distinct substrate-binding mode
Abstract
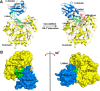
o-Succinylbenzoyl-CoA (OSB-CoA) synthetase (MenE) is an essential enzyme in bacterial vitamin K biosynthesis and an important target in the development of new antibiotics. It is a member of the adenylating enzymes (ANL) family, which reconfigure their active site in two different active conformations, one for the adenylation half-reaction and the other for a thioesterification half-reaction, in a domain-alternation catalytic mechanism. Although several aspects of the adenylating mechanism in MenE have recently been uncovered, its thioesterification conformation remains elusive. Here, using a catalytically competent Bacillus subtilis mutant protein complexed with an OSB-CoA analogue, we determined MenE high-resolution structures to 1.76 and 1.90 Å resolution in a thioester-forming conformation. By comparison with the adenylation conformation, we found that MenE's C-domain rotates around the Ser-384 hinge by 139.5° during domain-alternation catalysis. The structures also revealed a thioesterification active site specifically conserved among MenE orthologues and a substrate-binding mode distinct from those of many other acyl/aryl-CoA synthetases. Of note, using site-directed mutagenesis, we identified several residues that specifically contribute to the thioesterification half-reaction without affecting the adenylation half-reaction. Moreover, we observed a substantial movement of the activated succinyl group in the thioesterification half-reaction. These findings provide new insights into the domain-alternation catalysis of a bacterial enzyme essential for vitamin K biosynthesis and of its adenylating homologues in the ANL enzyme family.
Keywords: crystal structure; enzyme catalysis; enzyme mechanism; substrate specificity; vitamin K.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
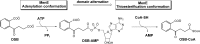



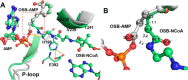
References
-
- Forsyth R. A., Haselbeck R. J., Ohlsen K. L., Yamamoto R. T., Xu H., Trawick J. D., Wall D., Wang L., Vickie B. D., Froelich J. M., Kedar G. C., King P., McCarthy M., Malone C., Misiner B., et al. (2002) A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43, 1387–1400 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

