Characterization of the scrambling domain of the TMEM16 family
- PMID: 28559311
- PMCID: PMC5474828
- DOI: 10.1073/pnas.1703391114
Characterization of the scrambling domain of the TMEM16 family
Abstract
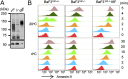
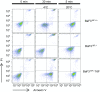
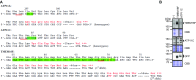
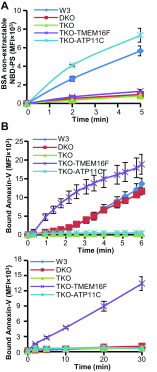
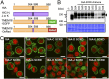
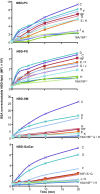
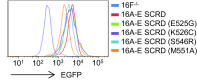
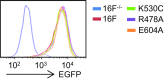
The TMEM16 protein family has 10 members, each of which carries 10 transmembrane segments. TMEM16A and 16B are Ca2+-activated Cl- channels. Several other members, including TMEM16F, promote phospholipid scrambling between the inner and outer leaflets of a cell membrane in response to intracellular Ca2+ However, the mechanism by which TMEM16 proteins translocate phospholipids in plasma membranes remains elusive. Here we show that Ca2+-activated, TMEM16F-supported phospholipid scrambling proceeds at 4 °C. Similar to TMEM16F and 16E, seven TMEM16 family members were found to carry a domain (SCRD; scrambling domain) spanning the fourth and fifth transmembrane segments that conferred scrambling ability to TMEM16A. By introducing point mutations into TMEM16F, we found that a lysine in the fourth transmembrane segment of the SCRD as well as an arginine in the third and a glutamic acid in the sixth transmembrane segment were important for exposing phosphatidylserine from the inner to the outer leaflet. However, their role in internalizing phospholipids was limited. Our results suggest that TMEM16 provides a cleft containing hydrophilic "stepping stones" for the outward translocation of phospholipids.
Keywords: TMEM16; phospholipids; point mutation; scramblase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Regulation of TMEM16A/ANO1 and TMEM16F/ANO6 ion currents and phospholipid scrambling by Ca2+ and plasma membrane lipid.J Physiol. 2018 Jan 15;596(2):217-229. doi: 10.1113/JP275175. Epub 2017 Dec 18. J Physiol. 2018. PMID: 29134661 Free PMC article.
-
Single-molecule analysis of phospholipid scrambling by TMEM16F.Proc Natl Acad Sci U S A. 2018 Mar 20;115(12):3066-3071. doi: 10.1073/pnas.1717956115. Epub 2018 Mar 5. Proc Natl Acad Sci U S A. 2018. PMID: 29507235 Free PMC article.
-
Calcium-dependent phospholipid scramblase activity of TMEM16 protein family members.J Biol Chem. 2013 May 10;288(19):13305-16. doi: 10.1074/jbc.M113.457937. Epub 2013 Mar 26. J Biol Chem. 2013. PMID: 23532839 Free PMC article.
-
Structural basis for phospholipid scrambling in the TMEM16 family.Curr Opin Struct Biol. 2016 Aug;39:61-70. doi: 10.1016/j.sbi.2016.05.020. Epub 2016 Jun 10. Curr Opin Struct Biol. 2016. PMID: 27295354 Review.
-
Established and emerging players in phospholipid scrambling: A structural perspective.Biochimie. 2024 Dec;227(Pt B):111-122. doi: 10.1016/j.biochi.2024.09.008. Epub 2024 Sep 18. Biochimie. 2024. PMID: 39304020 Review.
Cited by
-
The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes.Nat Struct Mol Biol. 2021 Oct;28(10):825-834. doi: 10.1038/s41594-021-00665-8. Epub 2021 Oct 8. Nat Struct Mol Biol. 2021. PMID: 34625749 Free PMC article.
-
Efferocytosis: The Janus-Faced Gatekeeper of Aging and Tumor Fate.Aging Cell. 2025 Feb;24(2):e14467. doi: 10.1111/acel.14467. Epub 2025 Jan 3. Aging Cell. 2025. PMID: 39748782 Free PMC article. Review.
-
The allosteric mechanism leading to an open-groove lipid conductive state of the TMEM16F scramblase.Commun Biol. 2022 Sep 19;5(1):990. doi: 10.1038/s42003-022-03930-8. Commun Biol. 2022. PMID: 36123525 Free PMC article.
-
Targeting lipid scrambling potentiates ferroptosis and triggers tumor immune rejection.Sci Adv. 2025 Aug 15;11(33):eadx6587. doi: 10.1126/sciadv.adx6587. Epub 2025 Aug 15. Sci Adv. 2025. PMID: 40815641 Free PMC article.
-
Structure-Function of TMEM16 Ion Channels and Lipid Scramblases.Adv Exp Med Biol. 2021;1349:87-109. doi: 10.1007/978-981-16-4254-8_6. Adv Exp Med Biol. 2021. PMID: 35138612 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

