A mechanism for lipid binding to apoE and the role of intrinsically disordered regions coupled to domain-domain interactions
- PMID: 28559318
- PMCID: PMC5474821
- DOI: 10.1073/pnas.1705080114
A mechanism for lipid binding to apoE and the role of intrinsically disordered regions coupled to domain-domain interactions
Abstract
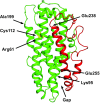
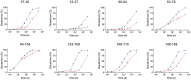
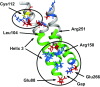
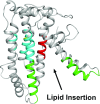
Relative to the apolipoprotein E (apoE) E3 allele of the APOE gene, apoE4 strongly increases the risk for the development of late-onset Alzheimer's disease. However, apoE4 differs from apoE3 by only a single amino acid at position 112, which is arginine in apoE4 and cysteine in apoE3. It remains unclear why apoE3 and apoE4 are functionally different. Described here is a proposal for understanding the functional differences between these two isoforms with respect to lipid binding. A mechanism is proposed that is based on the full-length monomeric structure of the protein, on hydrogen-deuterium exchange mass spectrometry data, and on the role of intrinsically disordered regions to control protein motions. It is proposed that lipid binds between the N-terminal and C-terminal domains and that separation of the two domains, along with the presence of intrinsically disordered regions, controls this process. The mechanism explains why apoE3 differs from apoE4 with respect to different lipid-binding specificities, why lipid increases the binding of apoE to its receptor, and why specific residues are conserved.
Keywords: apolipoprotein E; conserved residues; domain–domain interaction; hydrogen–deuterium exchange; protein structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Concerning the structure of apoE.Protein Sci. 2013 Dec;22(12):1820-5. doi: 10.1002/pro.2379. Epub 2013 Oct 19. Protein Sci. 2013. PMID: 24115173 Free PMC article.
-
Structural differences between apoE3 and apoE4 may be useful in developing therapeutic agents for Alzheimer's disease.Proc Natl Acad Sci U S A. 2012 Jun 5;109(23):8913-8. doi: 10.1073/pnas.1207022109. Epub 2012 May 21. Proc Natl Acad Sci U S A. 2012. PMID: 22615372 Free PMC article.
-
Molecular basis for the differences in lipid and lipoprotein binding properties of human apolipoproteins E3 and E4.Biochemistry. 2010 Dec 28;49(51):10881-9. doi: 10.1021/bi1017655. Epub 2010 Dec 3. Biochemistry. 2010. PMID: 21114327 Free PMC article.
-
Apolipoprotein E: from cardiovascular disease to neurodegenerative disorders.J Mol Med (Berl). 2016 Jul;94(7):739-46. doi: 10.1007/s00109-016-1427-y. Epub 2016 Jun 9. J Mol Med (Berl). 2016. PMID: 27277824 Free PMC article. Review.
-
Apolipoprotein E and oxidative stress in brain with relevance to Alzheimer's disease.Neurobiol Dis. 2020 May;138:104795. doi: 10.1016/j.nbd.2020.104795. Epub 2020 Feb 6. Neurobiol Dis. 2020. PMID: 32036033 Free PMC article. Review.
Cited by
-
MAD-microbial (origin of) Alzheimer's disease hypothesis: from infection and the antimicrobial response to disruption of key copper-based systems.Front Neurosci. 2024 Oct 2;18:1467333. doi: 10.3389/fnins.2024.1467333. eCollection 2024. Front Neurosci. 2024. PMID: 39416952 Free PMC article. Review.
-
Lipoproteins in the Central Nervous System: From Biology to Pathobiology.Annu Rev Biochem. 2022 Jun 21;91:731-759. doi: 10.1146/annurev-biochem-032620-104801. Epub 2022 Mar 18. Annu Rev Biochem. 2022. PMID: 35303786 Free PMC article. Review.
-
Endogenous Human Proteins Interfering with Amyloid Formation.Biomolecules. 2022 Mar 14;12(3):446. doi: 10.3390/biom12030446. Biomolecules. 2022. PMID: 35327638 Free PMC article. Review.
-
Molecular docking analysis of arjunolic acid from Terminalia arjuna with a coronary artery disease target APOE4.Bioinformation. 2021 Nov 30;17(11):949-958. doi: 10.6026/97320630017949. eCollection 2021. Bioinformation. 2021. PMID: 35655909 Free PMC article.
-
ApoE Lipidation as a Therapeutic Target in Alzheimer's Disease.Int J Mol Sci. 2020 Sep 1;21(17):6336. doi: 10.3390/ijms21176336. Int J Mol Sci. 2020. PMID: 32882843 Free PMC article. Review.
References
-
- Corder EH, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. - PubMed
-
- Saunders AM, et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology. 1993;43:1467–1472. - PubMed
-
- Dong LM, et al. Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia. Nat Struct Biol. 1996;3:718–722. - PubMed
-
- Phillips MC. Apolipoprotein E isoforms and lipoprotein metabolism. IUBMB Life. 2014;66:616–623. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

