Estimating the parameters of background selection and selective sweeps in Drosophila in the presence of gene conversion
- PMID: 28559322
- PMCID: PMC5474792
- DOI: 10.1073/pnas.1619434114
Estimating the parameters of background selection and selective sweeps in Drosophila in the presence of gene conversion
Abstract
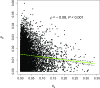
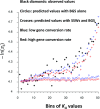
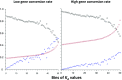
We used whole-genome resequencing data from a population of Drosophila melanogaster to investigate the causes of the negative correlation between the within-population synonymous nucleotide site diversity (πS ) of a gene and its degree of divergence from related species at nonsynonymous nucleotide sites (KA ). By using the estimated distributions of mutational effects on fitness at nonsynonymous and UTR sites, we predicted the effects of background selection at sites within a gene on πS and found that these could account for only part of the observed correlation between πS and KA We developed a model of the effects of selective sweeps that included gene conversion as well as crossing over. We used this model to estimate the average strength of selection on positively selected mutations in coding sequences and in UTRs, as well as the proportions of new mutations that are selectively advantageous. Genes with high levels of selective constraint on nonsynonymous sites were found to have lower strengths of positive selection and lower proportions of advantageous mutations than genes with low levels of constraint. Overall, background selection and selective sweeps within a typical gene reduce its synonymous diversity to ∼75% of its value in the absence of selection, with larger reductions for genes with high KA Gene conversion has a major effect on the estimates of the parameters of positive selection, such that the estimated strength of selection on favorable mutations is greatly reduced if it is ignored.
Keywords: Drosophila melanogaster; background selection; gene conversion; selective sweeps; sequence diversity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

