Validation of Suitable Reference Genes for RT-qPCR Data in Achyranthes bidentata Blume under Different Experimental Conditions
- PMID: 28559905
- PMCID: PMC5432617
- DOI: 10.3389/fpls.2017.00776
Validation of Suitable Reference Genes for RT-qPCR Data in Achyranthes bidentata Blume under Different Experimental Conditions
Abstract
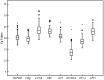
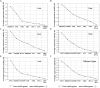
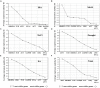
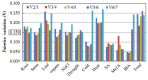
Real-time quantitative polymerase chain reaction (RT-qPCR) is a sensitive technique for gene expression studies. However, choosing the appropriate reference gene is essential to obtain reliable results for RT-qPCR assays. In the present work, the expression of eight candidate reference genes, EF1-α (elongation factor 1-α), GAPDH (glyceraldehyde 3-phosphate dehydrogenase), UBC (ubiquitin-conjugating enzyme), UBQ (polyubiquitin), ACT (actin), β-TUB (β-tubulin), APT1 (adenine phosphoribosyltransferase 1), and 18S rRNA (18S ribosomal RNA), was evaluated in Achyranthes bidentata samples using two algorithms, geNorm and NormFinder. The samples were classified into groups according to developmental stages, various tissues, stresses (cold, heat, drought, NaCl), and hormone treatments (MeJA, IBA, SA). Suitable combination of reference genes for RT-qPCR normalization should be applied according to different experimental conditions. In this study, EF1-α, UBC, and ACT genes were verified as the suitable reference genes across all tested samples. To validate the suitability of the reference genes, we evaluated the relative expression of CAS, which is a gene that may be involved in phytosterol synthesis. Our results provide the foundation for gene expression analysis in A. bidentata and other species of Amaranthaceae.
Keywords: Achyranthes bidentata Bl.; RT-qPCR data normalization; gene validation; reference genes; selection.
Figures


Similar articles
-
Validation of suitable reference genes for gene expression analysis in the halophyte Salicornia europaea by real-time quantitative PCR.Front Plant Sci. 2015 Jan 21;5:788. doi: 10.3389/fpls.2014.00788. eCollection 2014. Front Plant Sci. 2015. PMID: 25653658 Free PMC article.
-
Selection and validation of reliable reference genes for quantitative real-time PCR in Barnyard millet (Echinochloa spp.) under varied abiotic stress conditions.Sci Rep. 2023 Sep 20;13(1):15573. doi: 10.1038/s41598-023-40526-6. Sci Rep. 2023. PMID: 37731036 Free PMC article.
-
Identification and evaluation of reference genes for quantitative real-time PCR analysis in Polygonum cuspidatum based on transcriptome data.BMC Plant Biol. 2019 Nov 14;19(1):498. doi: 10.1186/s12870-019-2108-0. BMC Plant Biol. 2019. PMID: 31726985 Free PMC article.
-
Selection and validation of appropriate reference genes for real-time quantitative PCR analysis in Momordica charantia.Phytochemistry. 2019 Aug;164:1-11. doi: 10.1016/j.phytochem.2019.04.010. Epub 2019 May 2. Phytochemistry. 2019. PMID: 31054374
-
Current Practices for Reference Gene Selection in RT-qPCR of Aspergillus: Outlook and Recommendations for the Future.Genes (Basel). 2021 Jun 24;12(7):960. doi: 10.3390/genes12070960. Genes (Basel). 2021. PMID: 34202507 Free PMC article. Review.
Cited by
-
Selection and Validation of Appropriate Reference Genes for Quantitative RT-PCR Analysis in Rubia yunnanensis Diels Based on Transcriptome Data.Biomed Res Int. 2020 Jan 9;2020:5824841. doi: 10.1155/2020/5824841. eCollection 2020. Biomed Res Int. 2020. PMID: 31998793 Free PMC article.
-
Selection and Validation of Reference Genes for RT-qPCR Analysis in Spinacia oleracea under Abiotic Stress.Biomed Res Int. 2021 Feb 3;2021:4853632. doi: 10.1155/2021/4853632. eCollection 2021. Biomed Res Int. 2021. PMID: 33623781 Free PMC article.
-
Systematic Identification and Validation of Suitable Reference Genes for the Normalization of Gene Expression in Prunella vulgaris under Different Organs and Spike Development Stages.Genes (Basel). 2022 Oct 25;13(11):1947. doi: 10.3390/genes13111947. Genes (Basel). 2022. PMID: 36360184 Free PMC article.
-
Selection and evaluation of reference genes for qRT-PCR analysis in Euscaphis konishii Hayata based on transcriptome data.Plant Methods. 2018 Jun 4;14:42. doi: 10.1186/s13007-018-0311-x. eCollection 2018. Plant Methods. 2018. PMID: 29881443 Free PMC article.
-
Fungal endophyte-induced salidroside and tyrosol biosynthesis combined with signal cross-talk and the mechanism of enzyme gene expression in Rhodiola crenulata.Sci Rep. 2017 Oct 2;7(1):12540. doi: 10.1038/s41598-017-12895-2. Sci Rep. 2017. PMID: 28970519 Free PMC article.
References
-
- Andersen C. L., Jensen J. L., Ørntoft T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64 5245–5250. 10.1158/0008-5472.CAN-04-0496 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

