The gene family-free median of three
- PMID: 28559921
- PMCID: PMC5446766
- DOI: 10.1186/s13015-017-0106-z
The gene family-free median of three
Abstract
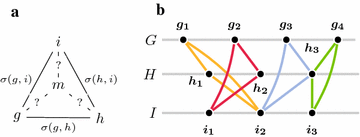
Background: The gene family-free framework for comparative genomics aims at providing methods for gene order analysis that do not require prior gene family assignment, but work directly on a sequence similarity graph. We study two problems related to the breakpoint median of three genomes, which asks for the construction of a fourth genome that minimizes the sum of breakpoint distances to the input genomes.
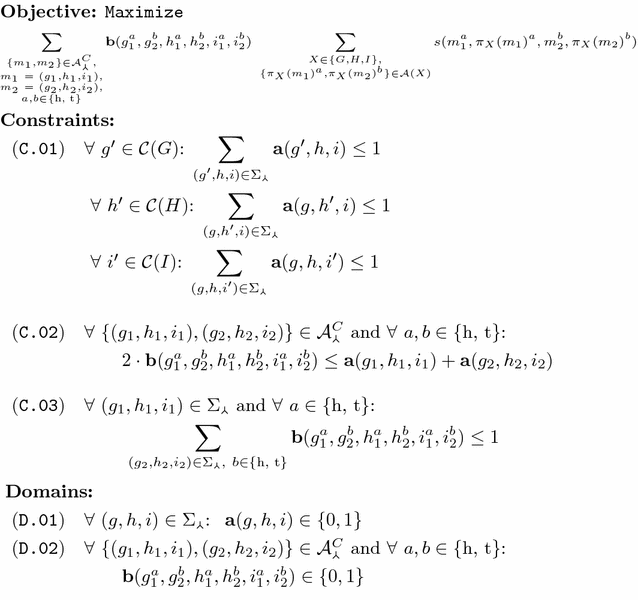
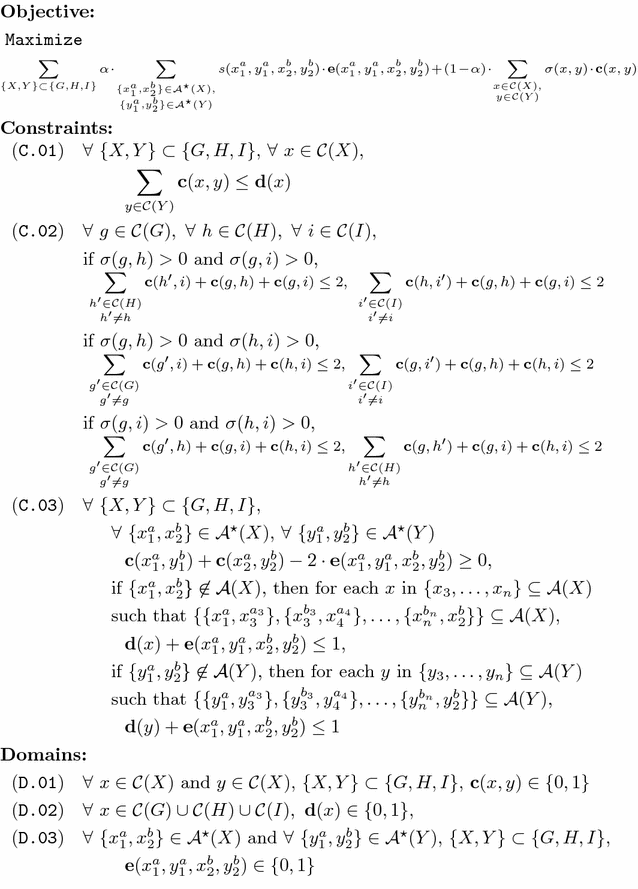
Methods: We present a model for constructing a median of three genomes in this family-free setting, based on maximizing an objective function that generalizes the classical breakpoint distance by integrating sequence similarity in the score of a gene adjacency. We study its computational complexity and we describe an integer linear program (ILP) for its exact solution. We further discuss a related problem called family-free adjacencies for k genomes for the special case of [Formula: see text] and present an ILP for its solution. However, for this problem, the computation of exact solutions remains intractable for sufficiently large instances. We then proceed to describe a heuristic method, FFAdj-AM, which performs well in practice.
Results: The developed methods compute accurate positional orthologs for genomes comparable in size of bacterial genomes on simulated data and genomic data acquired from the OMA orthology database. In particular, FFAdj-AM performs equally or better when compared to the well-established gene family prediction tool MultiMSOAR.
Conclusions: We study the computational complexity of a new family-free model and present algorithms for its solution. With FFAdj-AM, we propose an appealing alternative to established tools for identifying higher confidence positional orthologs.
Keywords: Breakpoint median; Family-free genome comparison; Positional orthology.
Figures



References
-
- Braga MDV, Chauve C, Doerr D, Jahn K, Stoye J, Thévenin A, Wittler R. The potential of family-free genome comparison. In: Models and algorithms for genome evolution. London: Springer; 2013. p. 287–323.
-
- Dörr D. Gene family-free genome comparison. PhD thesis, Universität Bielefeld, Bielefeld, Germany. 2016. https://pub.uni-bielefeld.de/publication/2902049.
LinkOut - more resources
Full Text Sources
Other Literature Sources

