Differentiation of Bifidobacterium longum subspecies longum and infantis by quantitative PCR using functional gene targets
- PMID: 28560114
- PMCID: PMC5446769
- DOI: 10.7717/peerj.3375
Differentiation of Bifidobacterium longum subspecies longum and infantis by quantitative PCR using functional gene targets
Abstract
Background: Members of the genus Bifidobacterium are abundant in the feces of babies during the exclusively-milk-diet period of life. Bifidobacterium longum is reported to be a common member of the infant fecal microbiota. However, B. longum is composed of three subspecies, two of which are represented in the bowel microbiota (B. longum subsp. longum; B. longum subsp. infantis). B. longum subspecies are not differentiated in many studies, so that their prevalence and relative abundances are not accurately known. This may largely be due to difficulty in assigning subspecies identity using DNA sequences of 16S rRNA or tuf genes that are commonly used in bacterial taxonomy.
Methods: We developed a qPCR method targeting the sialidase gene (subsp. infantis) and sugar kinase gene (subsp. longum) to differentiate the subspecies using specific primers and probes. Specificity of the primers/probes was tested by in silico, pangenomic search, and using DNA from standard cultures of bifidobacterial species. The utility of the method was further examined using DNA from feces that had been collected from infants inhabiting various geographical regions.
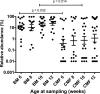
Results: A pangenomic search of the NCBI genomic database showed that the PCR primers/probes targeted only the respective genes of the two subspecies. The primers/probes showed total specificity when tested against DNA extracted from the gold standard strains (type cultures) of bifidobacterial species detected in infant feces. Use of the qPCR method with DNA extracted from the feces of infants of different ages, delivery method and nutrition, showed that subsp. infantis was detectable (0-32.4% prevalence) in the feces of Australian (n = 90), South-East Asian (n = 24), and Chinese babies (n = 91), but in all cases at low abundance (<0.01-4.6%) compared to subsp. longum (0.1-33.7% abundance; 21.4-100% prevalence).
Discussion: Our qPCR method differentiates B. longum subspecies longum and infantis using characteristic functional genes. It can be used as an identification aid for isolates of bifidobacteria, as well as in determining prevalence and abundance of the subspecies in feces. The method should thus be useful in ecological studies of the infant gut microbiota during early life where an understanding of the ecology of bifidobacterial species may be important in developing interventions to promote infant health.
Keywords: Bifidobacterium; Functional gene; Infantis; Longum; qPCR.
Conflict of interest statement
The authors declare there are no competing interests. Colin G. Prosser and Dianne Lowry are employees of Dairy Goat Cooperative (NZ) Ltd., Hamilton, New Zealand. Pheng Soon Lee and Khai Hong Wong are employees of Mead Johnson Nutrition, Marina Bay, Singapore.
Figures
References
-
- Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J, Jun W. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host and Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. - DOI - PubMed
-
- Biavati B, Castagnoli P, Crociani F, Trovatelli LD. Species of the bifidobacterium in the feces of infants. Microbiologica. 1984;7:341–345. - PubMed
-
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

