Selective analysis of cancer-cell intrinsic transcriptional traits defines novel clinically relevant subtypes of colorectal cancer
- PMID: 28561063
- PMCID: PMC5499209
- DOI: 10.1038/ncomms15107
Selective analysis of cancer-cell intrinsic transcriptional traits defines novel clinically relevant subtypes of colorectal cancer
Abstract
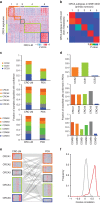
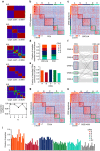
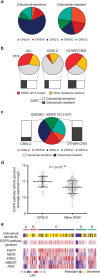
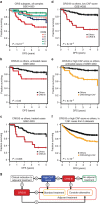
Stromal content heavily impacts the transcriptional classification of colorectal cancer (CRC), with clinical and biological implications. Lineage-dependent stromal transcriptional components could therefore dominate over more subtle expression traits inherent to cancer cells. Since in patient-derived xenografts (PDXs) stromal cells of the human tumour are substituted by murine counterparts, here we deploy human-specific expression profiling of CRC PDXs to assess cancer-cell intrinsic transcriptional features. Through this approach, we identify five CRC intrinsic subtypes (CRIS) endowed with distinctive molecular, functional and phenotypic peculiarities: (i) CRIS-A: mucinous, glycolytic, enriched for microsatellite instability or KRAS mutations; (ii) CRIS-B: TGF-β pathway activity, epithelial-mesenchymal transition, poor prognosis; (iii) CRIS-C: elevated EGFR signalling, sensitivity to EGFR inhibitors; (iv) CRIS-D: WNT activation, IGF2 gene overexpression and amplification; and (v) CRIS-E: Paneth cell-like phenotype, TP53 mutations. CRIS subtypes successfully categorize independent sets of primary and metastatic CRCs, with limited overlap on existing transcriptional classes and unprecedented predictive and prognostic performances.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- De Sousa E Melo F. et al.. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat. Med. 19, 614–618 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

