The Agassiz's desert tortoise genome provides a resource for the conservation of a threatened species
- PMID: 28562605
- PMCID: PMC5451010
- DOI: 10.1371/journal.pone.0177708
The Agassiz's desert tortoise genome provides a resource for the conservation of a threatened species
Abstract
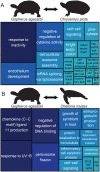
Agassiz's desert tortoise (Gopherus agassizii) is a long-lived species native to the Mojave Desert and is listed as threatened under the US Endangered Species Act. To aid conservation efforts for preserving the genetic diversity of this species, we generated a whole genome reference sequence with an annotation based on deep transcriptome sequences of adult skeletal muscle, lung, brain, and blood. The draft genome assembly for G. agassizii has a scaffold N50 length of 252 kbp and a total length of 2.4 Gbp. Genome annotation reveals 20,172 protein-coding genes in the G. agassizii assembly, and that gene structure is more similar to chicken than other turtles. We provide a series of comparative analyses demonstrating (1) that turtles are among the slowest-evolving genome-enabled reptiles, (2) amino acid changes in genes controlling desert tortoise traits such as shell development, longevity and osmoregulation, and (3) fixed variants across the Gopherus species complex in genes related to desert adaptations, including circadian rhythm and innate immune response. This G. agassizii genome reference and annotation is the first such resource for any tortoise, and will serve as a foundation for future analysis of the genetic basis of adaptations to the desert environment, allow for investigation into genomic factors affecting tortoise health, disease and longevity, and serve as a valuable resource for additional studies in this species complex.
Conflict of interest statement
Figures




Similar articles
-
Landscape limits gene flow and drives population structure in Agassiz's desert tortoise (Gopherus agassizii).Sci Rep. 2018 Jul 25;8(1):11231. doi: 10.1038/s41598-018-29395-6. Sci Rep. 2018. PMID: 30046050 Free PMC article.
-
Serologic and molecular evidence for Testudinid herpesvirus 2 infection in wild Agassiz's desert tortoises, Gopherus agassizii.J Wildl Dis. 2012 Jul;48(3):747-57. doi: 10.7589/0090-3558-48.3.747. J Wildl Dis. 2012. PMID: 22740541
-
Bilateral palpebral reduction and concurrent mycoplasmosis in a wild Agassiz's desert tortoise (Gopherus agassizii).Vet Ophthalmol. 2023 Jul;26(4):361-366. doi: 10.1111/vop.13089. Epub 2023 Apr 8. Vet Ophthalmol. 2023. PMID: 37030880
-
Mycoplasmosis and upper respiratory tract disease of tortoises: a review and update.Vet J. 2014 Sep;201(3):257-64. doi: 10.1016/j.tvjl.2014.05.039. Epub 2014 Jun 4. Vet J. 2014. PMID: 24951264 Review.
-
Role of urinary and cloacal bladders in chelonian water economy: historical and comparative perspectives.Biol Rev Camb Philos Soc. 1998 Nov;73(4):347-66. doi: 10.1017/s0006323198005210. Biol Rev Camb Philos Soc. 1998. PMID: 9951413 Review.
Cited by
-
Green, yellow or black? Genetic differentiation and adaptation signatures in a highly migratory marine turtle.Proc Biol Sci. 2021 Jul 14;288(1954):20210754. doi: 10.1098/rspb.2021.0754. Epub 2021 Jul 7. Proc Biol Sci. 2021. PMID: 34229490 Free PMC article.
-
The State of Squamate Genomics: Past, Present, and Future of Genome Research in the Most Speciose Terrestrial Vertebrate Order.Genes (Basel). 2023 Jul 1;14(7):1387. doi: 10.3390/genes14071387. Genes (Basel). 2023. PMID: 37510292 Free PMC article. Review.
-
A user's guide for understanding reptile and amphibian hydroregulation and climate change impacts.Conserv Physiol. 2025 Jun 16;13(1):coaf038. doi: 10.1093/conphys/coaf038. eCollection 2025. Conserv Physiol. 2025. PMID: 40575727 Free PMC article.
-
Complex immune responses and molecular reactions to pathogens and disease in a desert reptile (Gopherus agassizii).Ecol Evol. 2019 Feb 18;9(5):2516-2534. doi: 10.1002/ece3.4897. eCollection 2019 Mar. Ecol Evol. 2019. PMID: 30891197 Free PMC article.
-
Discovery of a New TLR Gene and Gene Expansion Event through Improved Desert Tortoise Genome Assembly with Chromosome-Scale Scaffolds.Genome Biol Evol. 2020 Feb 1;12(2):3917-3925. doi: 10.1093/gbe/evaa016. Genome Biol Evol. 2020. PMID: 32011707 Free PMC article.
References
-
- Franz R. The fossil record for North American tortoises In: Rostal DC, McCoy ED, Mushinsky HR, editors. Biology and Conservation of North American Tortoises. 2014. pp. 13–24.
-
- US Fish and Wildlife Service. Endangered and threatened wildlife and plants; determination of threatened status for the Mojave population of the desert tortoise. Federal Register. 1990; 55: 12178–12191.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

