Understanding RNA modifications: the promises and technological bottlenecks of the 'epitranscriptome'
- PMID: 28566301
- PMCID: PMC5451548
- DOI: 10.1098/rsob.170077
Understanding RNA modifications: the promises and technological bottlenecks of the 'epitranscriptome'
Abstract
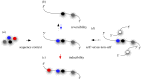
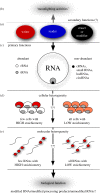
The discovery of mechanisms that alter genetic information via RNA editing or introducing covalent RNA modifications points towards a complexity in gene expression that challenges long-standing concepts. Understanding the biology of RNA modifications represents one of the next frontiers in molecular biology. To this date, over 130 different RNA modifications have been identified, and improved mass spectrometry approaches are still adding to this list. However, only recently has it been possible to map selected RNA modifications at single-nucleotide resolution, which has created a number of exciting hypotheses about the biological function of RNA modifications, culminating in the proposition of the 'epitranscriptome'. Here, we review some of the technological advances in this rapidly developing field, identify the conceptual challenges and discuss approaches that are needed to rigorously test the biological function of specific RNA modifications.
Keywords: RNA; chemical modification; epigenetics; transcriptome.
© 2017 The Authors.
Conflict of interest statement
We declare we have no competing interests.
Figures

References
-
- Machnicka MA, et al. 2013. MODOMICS: a database of RNA modification pathways—2013 update. Nucleic Acids Res. 41, D262–D267. (doi:10.1093/nar/gks1007) - DOI - PMC - PubMed
-
- Fersht AR. 1981. Enzymic editing mechanisms and the genetic code. Proc. R. Soc. Lond. B 212, 351–379. (doi:10.1098/rspb.1981.0044) - DOI - PubMed
-
- Benne R, Van den Burg J, Brakenhoff JP, Sloof P, Van Boom JH, Tromp MC. 1986. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46, 819–826. (doi:10.1016/0092-8674(86)90063-2) - DOI - PubMed
-
- Desrosiers R, Fridrich K, Rottmann F. 1974. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl Acad. Sci. USA 71, 3971 (doi:10.1073/pnas.71.10.3971) - DOI - PMC - PubMed
-
- Perry RP, Kelley DE, LaTorre J. 1974. Synthesis and turnover of nuclear and cytoplasmic polyadenylic acid in mouse L cells. J. Mol. Biol. 82, 315–331. (doi:10.1016/0022-2836(74)90593-2) - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

