Proof-of-Concept Gene Editing for the Murine Model of Inducible Arginase-1 Deficiency
- PMID: 28566761
- PMCID: PMC5451454
- DOI: 10.1038/s41598-017-02927-2
Proof-of-Concept Gene Editing for the Murine Model of Inducible Arginase-1 Deficiency
Abstract
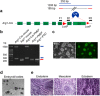
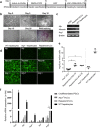
Arginase-1 deficiency in humans is a rare genetic disorder of metabolism resulting from a loss of arginase-1, leading to impaired ureagenesis, hyperargininemia and neurological deficits. Previously, we generated a tamoxifen-inducible arginase-1 deficient mouse model harboring a deletion of Arg1 exons 7 and 8 that leads to similar biochemical defects, along with a wasting phenotype and death within two weeks. Here, we report a strategy utilizing the Clustered, Regularly Interspaced, Short Palindromic Repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system in conjunction with piggyBac technology to target and reincorporate exons 7 and 8 at the specific Arg1 locus in attempts to restore the function of arginase-1 in induced pluripotent stem cell (iPSC)-derived hepatocyte-like cells (iHLCs) and macrophages in vitro. While successful gene targeted repair was achieved, minimal urea cycle function was observed in the targeted iHLCs compared to adult hepatocytes likely due to inadequate maturation of the cells. On the other hand, iPSC-derived macrophages expressed substantial amounts of "repaired" arginase. Our studies provide proof-of-concept for gene-editing at the Arg1 locus and highlight the challenges that lie ahead to restore sufficient liver-based urea cycle function in patients with urea cycle disorders.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

