Genomic prediction in early selection stages using multi-year data in a hybrid rye breeding program
- PMID: 28569139
- PMCID: PMC5452640
- DOI: 10.1186/s12863-017-0512-8
Genomic prediction in early selection stages using multi-year data in a hybrid rye breeding program
Abstract
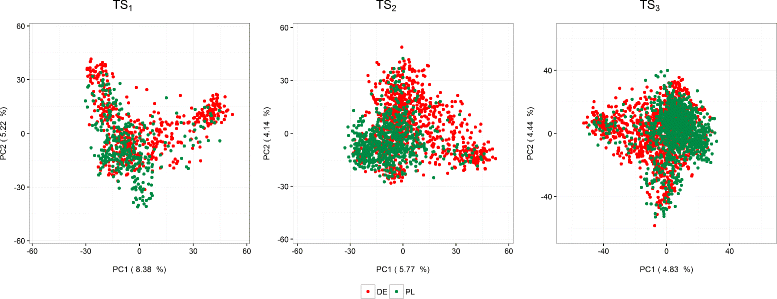
Background: The use of multiple genetic backgrounds across years is appealing for genomic prediction (GP) because past years' data provide valuable information on marker effects. Nonetheless, single-year GP models are less complex and computationally less demanding than multi-year GP models. In devising a suitable analysis strategy for multi-year data, we may exploit the fact that even if there is no replication of genotypes across years, there is plenty of replication at the level of marker loci. Our principal aim was to evaluate different GP approaches to simultaneously model genotype-by-year (GY) effects and breeding values using multi-year data in terms of predictive ability. The models were evaluated under different scenarios reflecting common practice in plant breeding programs, such as different degrees of relatedness between training and validation sets, and using a selected fraction of genotypes in the training set. We used empirical grain yield data of a rye hybrid breeding program. A detailed description of the prediction approaches highlighting the use of kinship for modeling GY is presented.
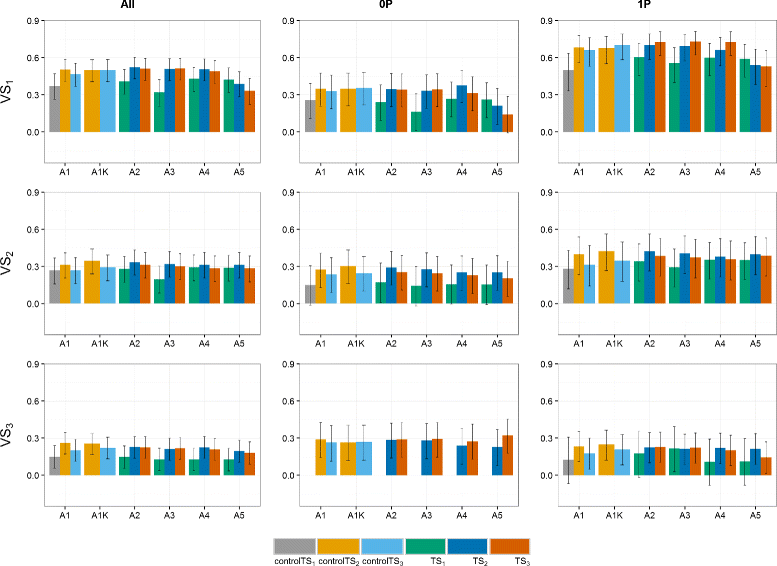
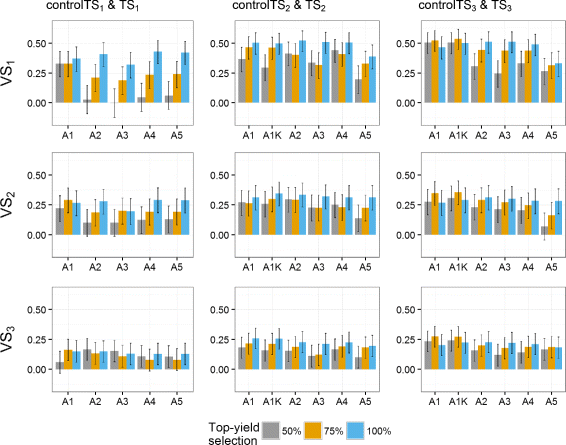
Results: Using the kinship to model GY was advantageous in particular for datasets disconnected across years. On average, predictive abilities were 5% higher for models using kinship to model GY over models without kinship. We confirmed that using data from multiple selection stages provides valuable GY information and helps increasing predictive ability. This increase is on average 30% higher when the predicted genotypes are closely related with the genotypes in the training set. A selection of top-yielding genotypes together with the use of kinship to model GY improves the predictive ability in datasets composed of single years of several selection cycles.
Conclusions: Our results clearly demonstrate that the use of multi-year data and appropriate modeling is beneficial for GP because it allows dissecting GY effects from genomic estimated breeding values. The model choice, as well as ensuring that the predicted candidates are sufficiently related to the genotypes in the training set, are crucial.
Keywords: Genomic prediction; Genotype-by-year interaction; Hybrid rye breeding; Multi-year data.
Figures





References
-
- Rutkoski J, Singh RP, Huerta-Espino J, Bhavani S, Poland J, Jannink JL, Sorrells ME. Efficient use of historical data for genomic selection: A case study of stem rust resistance in wheat. Plant Genome. 2015;8(1). - PubMed
-
- Schulz-Streeck T, Ogutu JO, Karaman Z, Knaak C, Piepho HP. Genomic selection using multiple populations. Crop Sci. 2012;52:2453–61. doi: 10.2135/cropsci2012.03.0160. - DOI
-
- Auinger HJ, Schönleben M, Lehermeier C, Schmidt M, Korzun V, Geiger HH, Piepho HP, Gordillo A, Wilde P, Bauer E, Schön CC. Model training across multiple breeding cycles significantly improves genomic prediction accuracy in rye (Secale cereale L) Theor Appl Genet. 2016;129:2043–53. doi: 10.1007/s00122-016-2756-5. - DOI - PMC - PubMed
-
- Schmidt M, Kollers S, Maasberg-Prelle A, Großer J, Schinkel B, Tomerius A, Graner A, Korzun V. Prediction of malting quality traits in barley based on genome-wide marker data to assess the potential of genomic selection. Theor Appl Genet. 2016;129:203–13. doi: 10.1007/s00122-015-2639-1. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

