The developmental regulator PKL is required to maintain correct DNA methylation patterns at RNA-directed DNA methylation loci
- PMID: 28569170
- PMCID: PMC5452414
- DOI: 10.1186/s13059-017-1226-y
The developmental regulator PKL is required to maintain correct DNA methylation patterns at RNA-directed DNA methylation loci
Abstract
Background: The chromodomain helicase DNA-binding family of ATP-dependent chromatin remodeling factors play essential roles during eukaryote growth and development. They are recruited by specific transcription factors and regulate the expression of developmentally important genes. Here, we describe an unexpected role in non-coding RNA-directed DNA methylation in Arabidopsis thaliana.
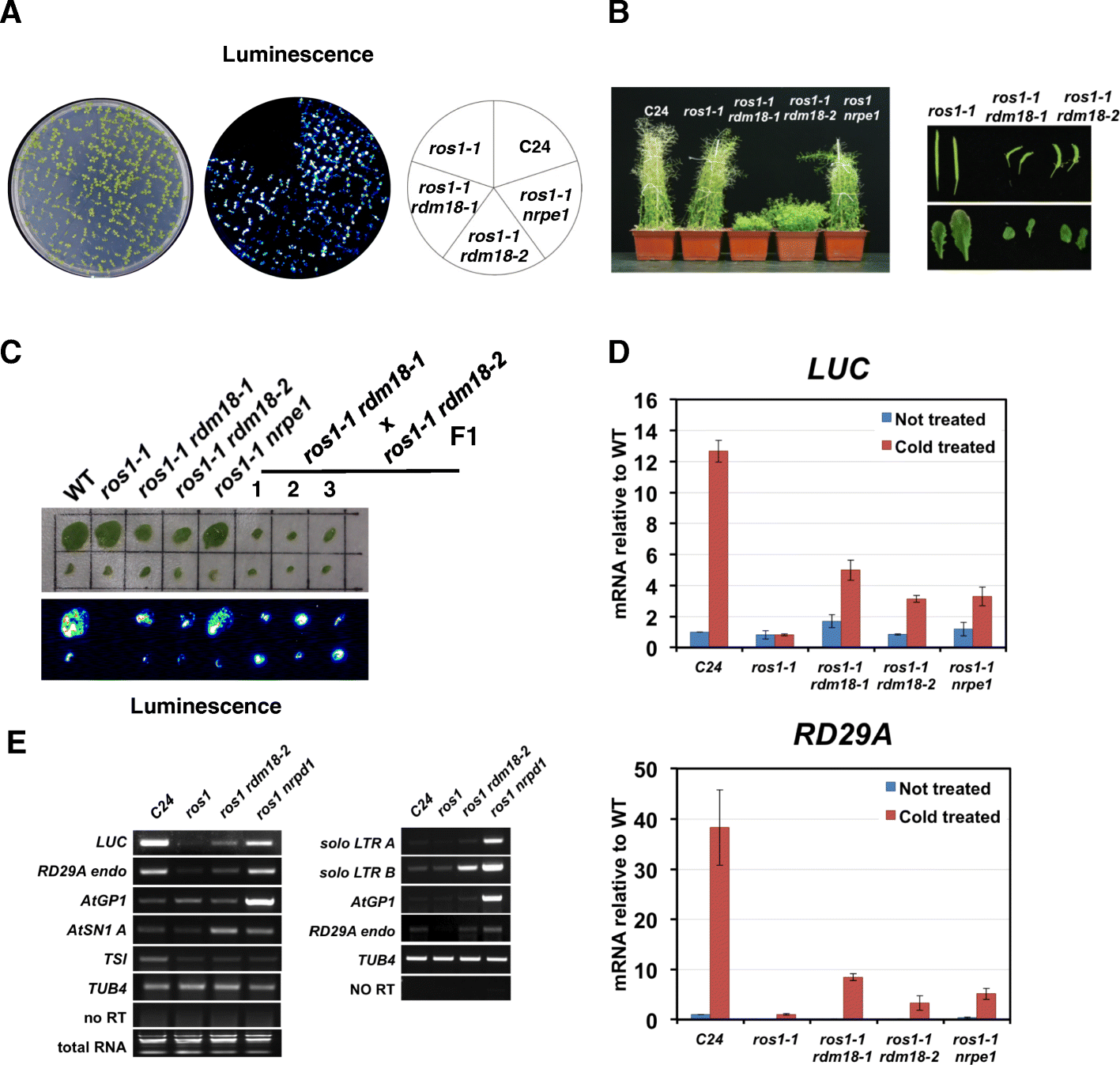
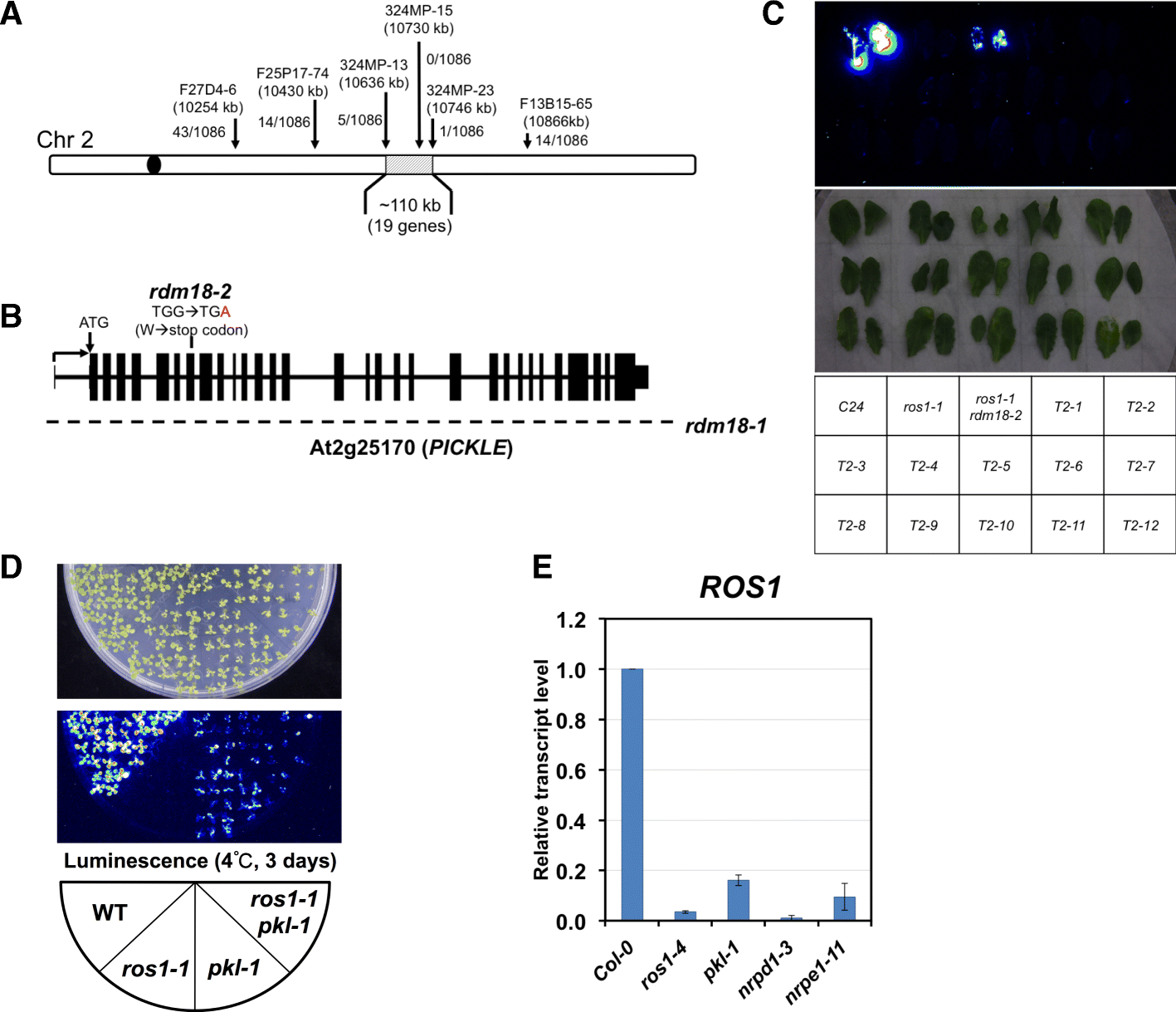
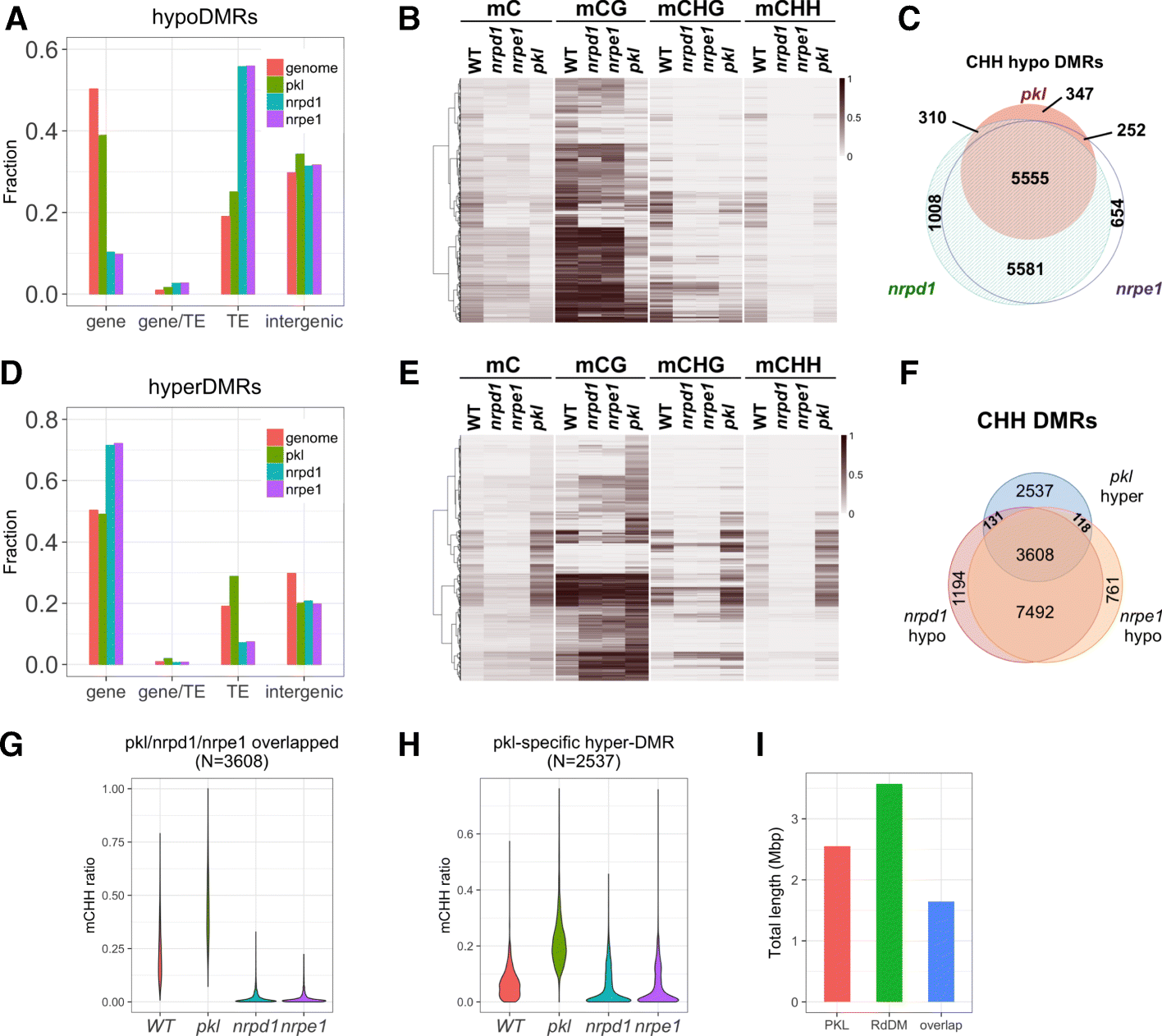
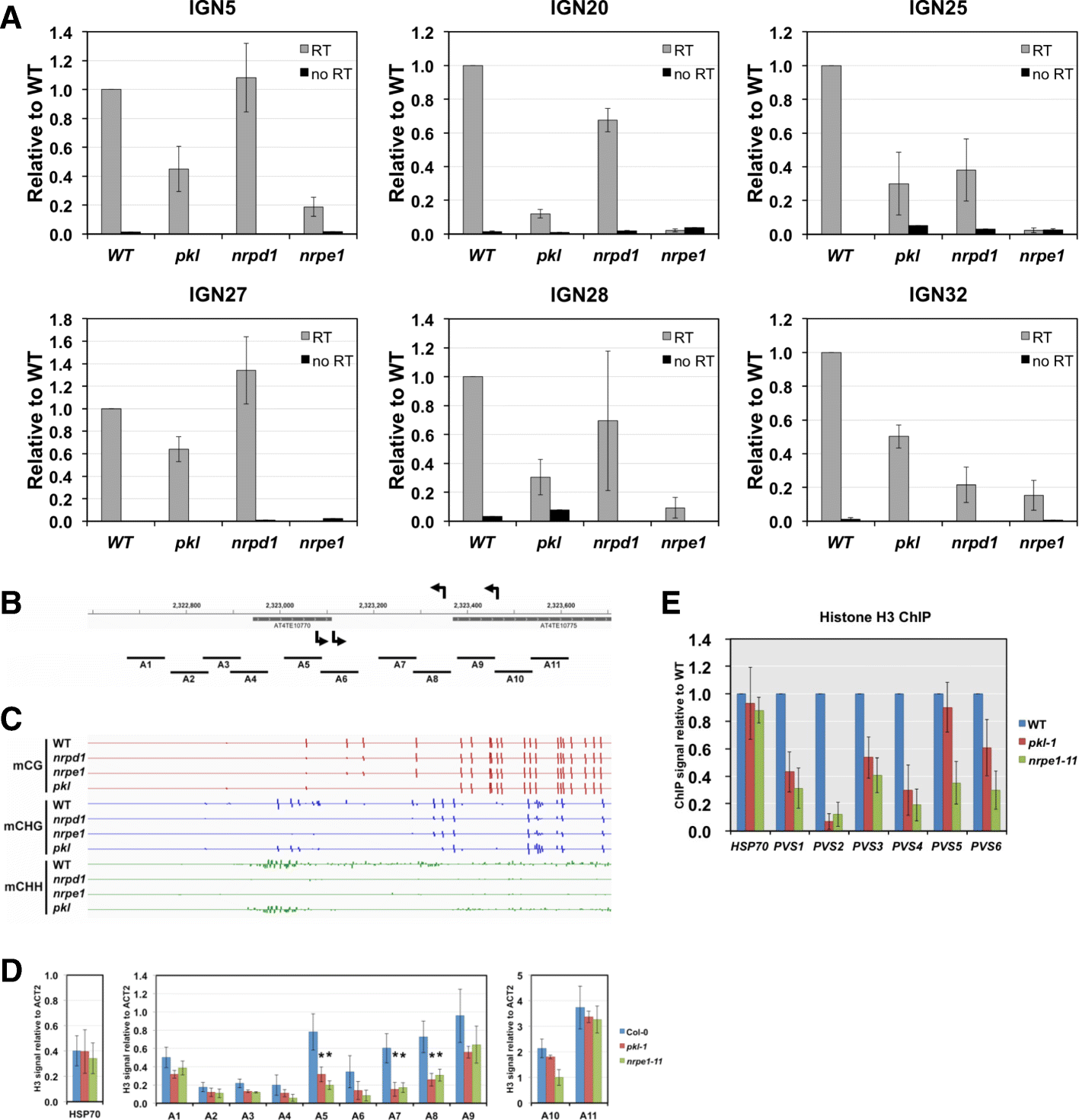
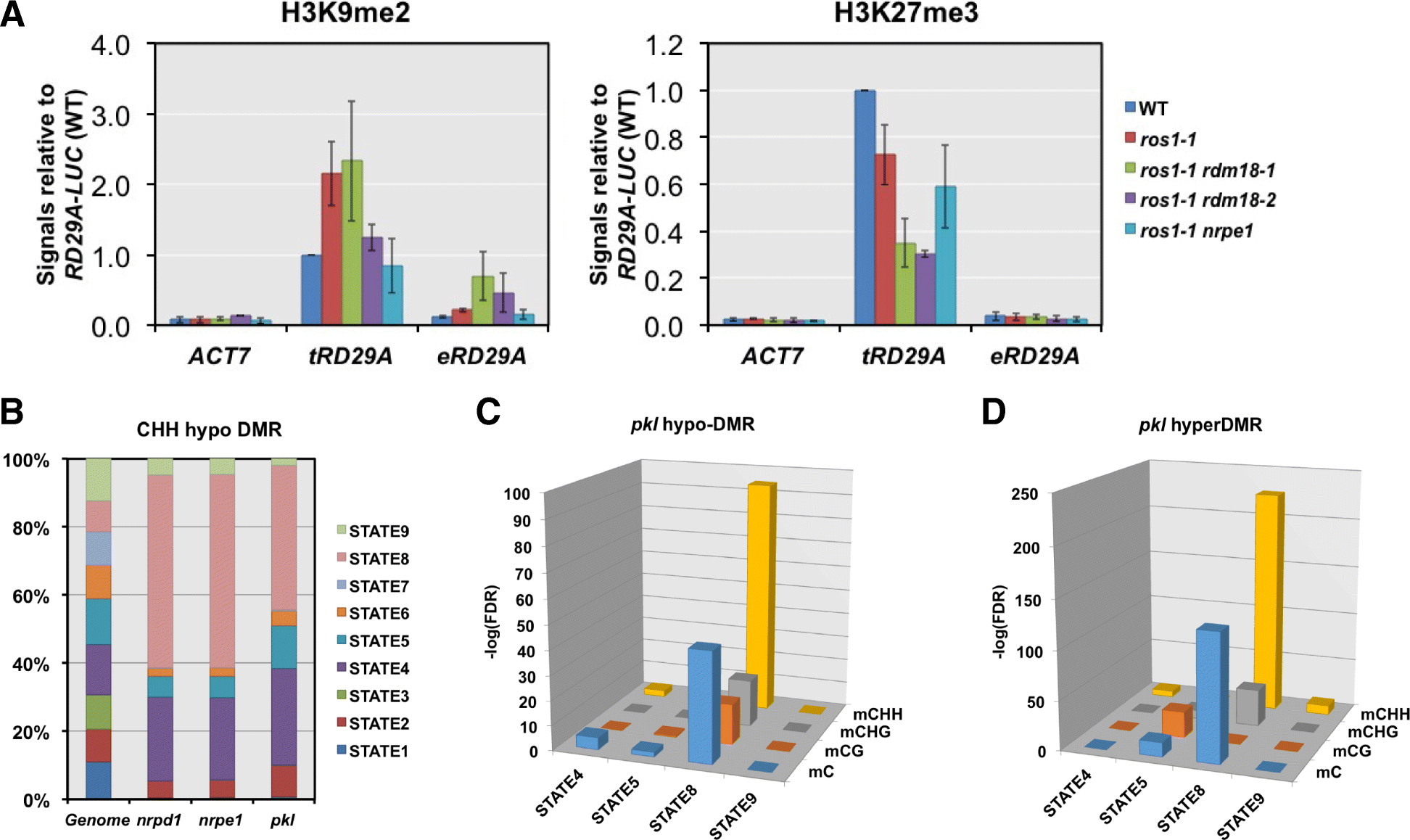
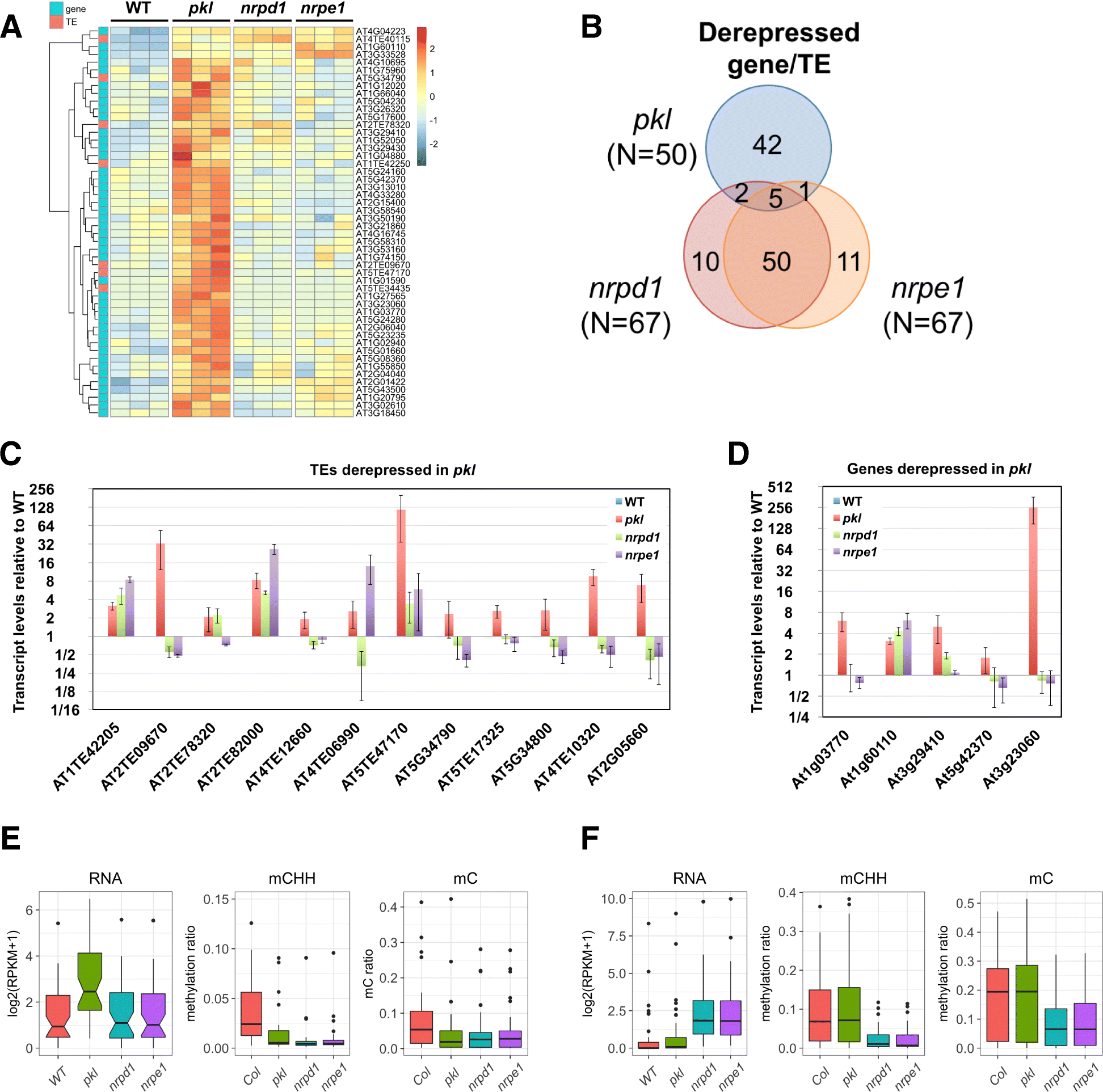
Results: Through forward genetic screens we identified PKL, a gene required for developmental regulation in plants, as a factor promoting transcriptional silencing at the transgenic RD29A promoter. Mutation of PKL results in DNA methylation changes at more than half of the loci that are targeted by RNA-directed DNA methylation (RdDM). A small number of transposable elements and genes had reduced DNA methylation correlated with derepression in the pkl mutant, though for the majority, decreases in DNA methylation are not sufficient to cause release of silencing. The changes in DNA methylation in the pkl mutant are positively correlated with changes in 24-nt siRNA levels. In addition, PKL is required for the accumulation of Pol V-dependent transcripts and for the positioning of Pol V-stabilized nucleosomes at several tested loci, indicating that RNA polymerase V-related functions are impaired in the pkl mutant.
Conclusions: PKL is required for transcriptional silencing and has significant effects on RdDM in plants. The changes in DNA methylation in the pkl mutant are correlated with changes in the non-coding RNAs produced by Pol IV and Pol V. We propose that at RdDM target regions, PKL may be required to create a chromatin environment that influences non-coding RNA production, DNA methylation, and transcriptional silencing.
Keywords: ATP-dependent chromatin remodeling; DNA methylation; Non-coding RNA (ncRNA); RNA-directed DNA methylation (RdDM).
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

