Sources of discordance among germ-line variant classifications in ClinVar
- PMID: 28569743
- PMCID: PMC5632819
- DOI: 10.1038/gim.2017.60
Sources of discordance among germ-line variant classifications in ClinVar
Erratum in
-
Corrigendum: Sources of discordance among germ-line variant classifications in ClinVar.Genet Med. 2018 Feb;20(2):282. doi: 10.1038/gim.2017.198. Epub 2017 Dec 7. Genet Med. 2018. PMID: 29215652 Free PMC article.
Abstract
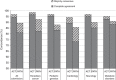
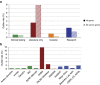
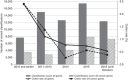
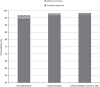
PurposeClinVar is increasingly used as a resource for both genetic variant interpretation and clinical practice. However, controversies exist regarding the consistency of classifications in ClinVar, and questions remain about how best to use these data. Our study systematically examined ClinVar to identify common sources of discordance and thus inform ongoing practices.MethodsWe analyzed variants that had multiple classifications in ClinVar, excluding benign polymorphisms. Classifications were categorized by potential actionability and pathogenicity. Consensus interpretations were calculated for each variant, and the properties of the discordant outlier classifications were summarized.ResultsOur study included 74,065 classifications of 27,224 unique variants in 1,713 genes. We found that (i) concordance rates differed among clinical areas and variant types; (ii) clinical testing methods had much higher concordance than basic literature curation and research efforts; (iii) older classifications had greater discordance than newer ones; and (iv) low-penetrance variants had particularly high discordance.ConclusionRecent variant classifications from clinical testing laboratories have high overall concordance in many (but not all) clinical areas. ClinVar can be a reliable resource supporting variant interpretation, quality assessment, and clinical practice when factors uncovered in this study are taken into account. Ongoing improvements to ClinVar may make it easier to use, particularly for nonexpert users.
Conflict of interest statement
All authors are employees of Invitae, a laboratory offering clinical genetic testing services. This project was funded by Invitae.
Figures

References
-
- National Institutes of Health Genetic Testing Registryhttp://www.ncbi.nlm.nih.gov/gtr/. Accessed 12 January 2017.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

