Deletion of a mycobacterial divisome factor collapses single-cell phenotypic heterogeneity
- PMID: 28569798
- PMCID: PMC5567998
- DOI: 10.1038/nature22361
Deletion of a mycobacterial divisome factor collapses single-cell phenotypic heterogeneity
Abstract
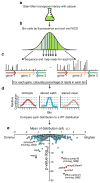
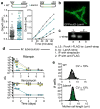
Microorganisms are often studied as populations but the behaviour of single, individual cells can have important consequences. For example, tuberculosis, caused by the bacterial pathogen Mycobacterium tuberculosis, requires months of antibiotic therapy even though the bulk of the bacterial population dies rapidly. Shorter courses lead to high rates of relapse because subpopulations of bacilli can survive despite being genetically identical to those that are easily killed. In fact, mycobacteria create variability each time a cell divides, producing daughter cells with different sizes and growth rates. The mechanism(s) that underlie this high-frequency variation and how variability relates to survival of the population are unknown. Here we show that mycobacteria actively create heterogeneity. Using a fluorescent reporter and a fluorescence-activated cell sorting (FACS)-based transposon screen, we find that deletion of lamA, a gene of previously unknown function, decreases heterogeneity in the population by decreasing asymmetric polar growth. LamA has no known homologues in other organisms, but is highly conserved across mycobacterial species. We find that LamA is a member of the mycobacterial division complex (the 'divisome'). It inhibits growth at nascent new poles, creating asymmetry in polar growth. The kinetics of killing individual cells that lack lamA are more uniform and more rapid with rifampicin and drugs that target the cell wall. Our results show that mycobacteria encode a non-conserved protein that controls the pattern of cell growth, resulting in a population that is both heterogeneous and better able to survive antibiotic pressure.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
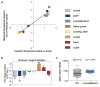
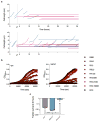



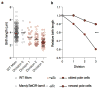

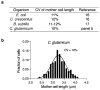



Comment in
-
Microbiology: Diversity breeds tolerance.Nature. 2017 May 31;546(7656):44-45. doi: 10.1038/546044a. Nature. 2017. PMID: 28569816 No abstract available.
References
-
- Mitchison DA. Role of individual drugs in the chemotherapy of tuberculosis. The International Journal of Tuberculosis and Lung Disease. 2000;4:796–806. - PubMed
-
- Santi I, Dhar N, Bousbaine D, Wakamoto Y, McKinney JD. Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria. Nature communications. 2013;4:2470. - PubMed
-
- Tiberghien F, Loor F. Ranking of P-glycoprotein substrates and inhibitors by a calcein-AM fluorometry screening assay. Anti-Cancer Drugs. 1996;7:568–578. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

