Characterization of DNA Binding by the Isolated N-Terminal Domain of Vaccinia Virus DNA Topoisomerase IB
- PMID: 28570045
- PMCID: PMC7251655
- DOI: 10.1021/acs.biochem.7b00042
Characterization of DNA Binding by the Isolated N-Terminal Domain of Vaccinia Virus DNA Topoisomerase IB
Abstract
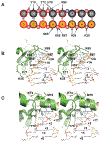
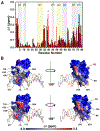
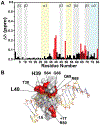
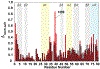
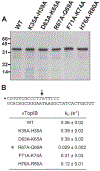
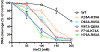
Vaccinia TopIB (vTopIB), a 314-amino acid eukaryal-type IB topoisomerase, recognizes and transesterifies at the DNA sequence 5'-(T/C)CCTT↓, leading to the formation of a covalent DNA-(3'-phosphotyrosyl274)-enzyme intermediate in the supercoil relaxation reaction. The C-terminal segment of vTopIB (amino acids 81-314), which engages the DNA minor groove at the scissile phosphodiester, comprises an autonomous catalytic domain that retains cleavage specificity, albeit with a cleavage site affinity lower than that of the full-length enzyme. The N-terminal domain (amino acids 1-80) engages the major groove on the DNA face opposite the scissile phosphodiester. Whereas DNA contacts of the N-terminal domain have been implicated in the DNA site affinity of vTopIB, it was not known whether the N-terminal domain per se could bind DNA. Here, using isothermal titration calorimetry, we demonstrate the ability of the isolated N-terminal domain to bind a CCCTT-containing 24-mer duplex with an apparent affinity that is ∼2.2-fold higher than that for an otherwise identical duplex in which the pentapyrimidine sequence is changed to ACGTG. Analyses of the interactions of the isolated N-terminal domain with duplex DNA via solution nuclear magnetic resonance methods are consistent with its DNA contacts observed in DNA-bound crystal structures of full-length vTopIB. The chemical shift perturbations and changes in hydrodynamic properties triggered by CCCTT DNA versus non-CCCTT DNA suggest differences in DNA binding dynamics. The importance of key N-terminal domain contacts in the context of full-length vTopIB is underscored by assessing the effects of double-alanine mutations on DNA transesterification and its sensitivity to ionic strength.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Similar articles
-
Chemical and traditional mutagenesis of vaccinia DNA topoisomerase provides insights to cleavage site recognition and transesterification chemistry.J Biol Chem. 2008 Jun 6;283(23):16093-103. doi: 10.1074/jbc.M801595200. Epub 2008 Mar 25. J Biol Chem. 2008. PMID: 18367446 Free PMC article.
-
Site-specific DNA transesterification by vaccinia topoisomerase: role of specific phosphates and nucleosides.Biochemistry. 1999 Dec 14;38(50):16599-612. doi: 10.1021/bi992001d. Biochemistry. 1999. PMID: 10600122
-
Mutational analysis of 26 residues of vaccinia DNA topoisomerase identifies Ser-204 as important for DNA binding and cleavage.Biochemistry. 1997 Jul 1;36(26):7944-50. doi: 10.1021/bi970498q. Biochemistry. 1997. PMID: 9201940
-
Vaccinia virus DNA topoisomerase: a model eukaryotic type IB enzyme.Biochim Biophys Acta. 1998 Oct 1;1400(1-3):321-37. doi: 10.1016/s0167-4781(98)00144-4. Biochim Biophys Acta. 1998. PMID: 9748643 Review.
-
Domains of human topoisomerase I and associated functions.Prog Nucleic Acid Res Mol Biol. 1998;60:111-32. doi: 10.1016/s0079-6603(08)60891-0. Prog Nucleic Acid Res Mol Biol. 1998. PMID: 9594573 Review.
Cited by
-
Irinotecan-Still an Important Player in Cancer Chemotherapy: A Comprehensive Overview.Int J Mol Sci. 2020 Jul 12;21(14):4919. doi: 10.3390/ijms21144919. Int J Mol Sci. 2020. PMID: 32664667 Free PMC article. Review.
-
Single-Cell Transcriptomic Changes in Patient-Derived Glioma and U87 Glioblastoma Cell Cultures Infected with the Oncolytic Virus VV-GMCSF-Lact.Int J Mol Sci. 2025 Jul 20;26(14):6983. doi: 10.3390/ijms26146983. Int J Mol Sci. 2025. PMID: 40725231 Free PMC article.
-
Twist and Turn-Topoisomerase Functions in Mitochondrial DNA Maintenance.Int J Mol Sci. 2019 Apr 25;20(8):2041. doi: 10.3390/ijms20082041. Int J Mol Sci. 2019. PMID: 31027213 Free PMC article. Review.
-
Potential Inhibitors of Monkeypox Virus Revealed by Molecular Modeling Approach to Viral DNA Topoisomerase I.Molecules. 2023 Feb 2;28(3):1444. doi: 10.3390/molecules28031444. Molecules. 2023. PMID: 36771105 Free PMC article.
References
-
- Leppard JB, and Champoux JJ (2005) Human DNA topoisomerase I: relaxation, roles, and damage control, Chromosoma 114, 75–85. - PubMed
-
- Cheng C, Kussie P, Pavletich N, and Shuman S (1998) Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases, Cell 92, 841–850. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

