A review of active learning approaches to experimental design for uncovering biological networks
- PMID: 28570593
- PMCID: PMC5453429
- DOI: 10.1371/journal.pcbi.1005466
A review of active learning approaches to experimental design for uncovering biological networks
Abstract
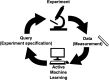
Various types of biological knowledge describe networks of interactions among elementary entities. For example, transcriptional regulatory networks consist of interactions among proteins and genes. Current knowledge about the exact structure of such networks is highly incomplete, and laboratory experiments that manipulate the entities involved are conducted to test hypotheses about these networks. In recent years, various automated approaches to experiment selection have been proposed. Many of these approaches can be characterized as active machine learning algorithms. Active learning is an iterative process in which a model is learned from data, hypotheses are generated from the model to propose informative experiments, and the experiments yield new data that is used to update the model. This review describes the various models, experiment selection strategies, validation techniques, and successful applications described in the literature; highlights common themes and notable distinctions among methods; and identifies likely directions of future research and open problems in the area.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Ideker TE, Thorsson V, Karp RM. Discovery of regulatory interactions through perturbation: inference and experimental design. In: Pacific Symposium on Biocomputing. vol. 5; 2000. p. 302–313. - PubMed
-
- Yeang CH, Mak HC, McCuine S, Workman C, Jaakkola T, Ideker T. Validation and refinement of gene-regulatory pathways on a network of physical interactions. Genome Biology. 2005;6(7):R62 doi: 10.1186/gb-2005-6-7-r62 - DOI - PMC - PubMed
-
- Pournara I, Wernisch L. Reconstruction of gene networks using Bayesian learning and manipulation experiments. Bioinformatics. 2004;20(17):2934–42. doi: 10.1093/bioinformatics/bth337 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

