Designing small molecules to target cryptic pockets yields both positive and negative allosteric modulators
- PMID: 28570708
- PMCID: PMC5453556
- DOI: 10.1371/journal.pone.0178678
Designing small molecules to target cryptic pockets yields both positive and negative allosteric modulators
Abstract
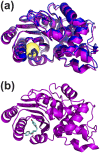
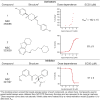
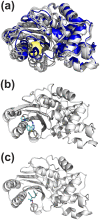
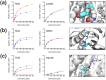
Allosteric drugs, which bind to proteins in regions other than their main ligand-binding or active sites, make it possible to target proteins considered "undruggable" and to develop new therapies that circumvent existing resistance. Despite growing interest in allosteric drug discovery, rational design is limited by a lack of sufficient structural information about alternative binding sites in proteins. Previously, we used Markov State Models (MSMs) to identify such "cryptic pockets," and here we describe a method for identifying compounds that bind in these cryptic pockets and modulate enzyme activity. Experimental tests validate our approach by revealing both an inhibitor and two activators of TEM β-lactamase (TEM). To identify hits, a library of compounds is first virtually screened against either the crystal structure of a known cryptic pocket or an ensemble of structures containing the same cryptic pocket that is extracted from an MSM. Hit compounds are then screened experimentally and characterized kinetically in individual assays. We identify three hits, one inhibitor and two activators, demonstrating that screening for binding to allosteric sites can result in both positive and negative modulation. The hit compounds have modest effects on TEM activity, but all have higher affinities than previously identified inhibitors, which bind the same cryptic pocket but were found, by chance, via a computational screen targeting the active site. Site-directed mutagenesis of key contact residues predicted by the docking models is used to confirm that the compounds bind in the cryptic pocket as intended. Because hit compounds are identified from docking against both the crystal structure and structures from the MSM, this platform should prove suitable for many proteins, particularly targets whose crystal structures lack obvious druggable pockets, and for identifying both inhibitory and activating small-molecule modulators.
Conflict of interest statement
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

