Domain alternation and active site remodeling are conserved structural features of ubiquitin E1
- PMID: 28572513
- PMCID: PMC5519361
- DOI: 10.1074/jbc.M117.787622
Domain alternation and active site remodeling are conserved structural features of ubiquitin E1
Abstract
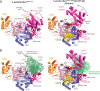
E1 enzymes for ubiquitin (Ub) and Ub-like modifiers (Ubls) harbor two catalytic activities that are required for Ub/Ubl activation: adenylation and thioester bond formation. Structural studies of the E1 for the Ubl
Keywords: E1; X-ray crystallography; adenylation; conformational change; domain alternation; enzyme mechanism; enzyme structure; structure-function; thioester; ubiquitin.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Komander D., and Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 - PubMed
-
- van der Veen A. G., and Ploegh H. L. (2012) Ubiquitin-like proteins. Annu. Rev. Biochem. 81, 323–357 - PubMed
-
- Haas A. L., and Rose I. A. (1982) The mechanism of ubiquitin activating enzyme. A kinetic and equilibrium analysis. J. Biol. Chem. 257, 10329–10337 - PubMed
-
- Haas A. L., Warms J. V., Hershko A., and Rose I. A. (1982) Ubiquitin-activating enzyme. Mechanism and role in protein-ubiquitin conjugation. J. Biol. Chem. 257, 2543–2548 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

