Overexpression of the human antigen R suppresses the immediate paradoxical proliferation of melanoma cell subpopulations in response to suboptimal BRAF inhibition
- PMID: 28573821
- PMCID: PMC5504324
- DOI: 10.1002/cam4.1091
Overexpression of the human antigen R suppresses the immediate paradoxical proliferation of melanoma cell subpopulations in response to suboptimal BRAF inhibition
Abstract
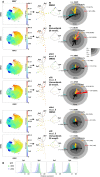
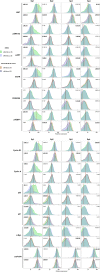
Tumor plasticity and the heterogeneous response of melanoma cells to targeted therapies are major limits for the long-term efficacy of this line of therapy. Targeting tumor plasticity is theoretically possible through the modulation of the expression of RNA-binding proteins which can affect many different compensatory mechanisms of the adaptive response of malignant cells to targeted therapies. Human antigen R (HuR) is a modulator of gene expression and a transacting factor in the mRNA-processing machinery used in the cell stress response, and is a potential target for reducing tumor plasticity. In this experiment, we exploit the inherent heterogeneous response of the A375 melanoma line to suboptimal BRAF inhibition as a model of immediate adaptive response. We first observe that HuR overexpression can prevent the heterogeneous response and thus the immediate paradoxical proliferation induced by low-doses vemurafenib treatment. We then use single-cell mass cytometry to characterize subpopulations, including those that paradoxically proliferate, based on their proliferation rate and the expression patterns of markers involved in the reversible adaptive resistance to BRAF inhibition and/or recognized as HuR targets involved in cell cycle regulation. Under suboptimal BRAF inhibition, HuR overexpression affects these subpopulations and their expression pattern with contrasting responses depending on their proliferation rate: faster-proliferating vemurafenib-sensitive or -resistant subpopulations showed higher death tendency and reduced size, and slower-proliferating subpopulations showed an attenuated resistant expression response and their paradoxical proliferation was inhibited. These observations pave the way to new therapeutic strategies for preventing the heterogeneous response of tumors to targeted therapies.
Keywords: BRAF inhibitor; RNA-binding protein HuR; cell heterogeneity; melanoma; single-cell mass cytometry; targeted therapy.
© 2017 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
Figures

Similar articles
-
Drug-induced expression of the RNA-binding protein HuR attenuates the adaptive response to BRAF inhibition in melanoma.Biochem Biophys Res Commun. 2019 Sep 17;517(2):181-187. doi: 10.1016/j.bbrc.2019.06.154. Epub 2019 Jul 3. Biochem Biophys Res Commun. 2019. PMID: 31279529
-
BET and BRAF inhibitors act synergistically against BRAF-mutant melanoma.Cancer Med. 2016 Jun;5(6):1183-93. doi: 10.1002/cam4.667. Epub 2016 May 11. Cancer Med. 2016. PMID: 27169980 Free PMC article.
-
Overcoming acquired BRAF inhibitor resistance in melanoma via targeted inhibition of Hsp90 with ganetespib.Mol Cancer Ther. 2014 Feb;13(2):353-63. doi: 10.1158/1535-7163.MCT-13-0481. Epub 2014 Jan 7. Mol Cancer Ther. 2014. PMID: 24398428
-
BRAF as a target for cancer therapy.Anticancer Agents Med Chem. 2011 Mar;11(3):285-95. doi: 10.2174/187152011795347469. Anticancer Agents Med Chem. 2011. PMID: 21426297 Review.
-
Immune Responses to BRAF-Targeted Therapy in Melanoma: Is Targeted Therapy Immunotherapy?Crit Rev Oncog. 2016;21(1-2):83-91. doi: 10.1615/CritRevOncog.2016017150. Crit Rev Oncog. 2016. PMID: 27481005 Review.
Cited by
-
Beyond the message: advantages of snapshot proteomics with single-cell mass cytometry in solid tumors.FEBS J. 2019 Apr;286(8):1523-1539. doi: 10.1111/febs.14730. Epub 2019 Jan 7. FEBS J. 2019. PMID: 30549207 Free PMC article. Review.
-
Mechanisms of Acquired BRAF Inhibitor Resistance in Melanoma: A Systematic Review.Cancers (Basel). 2020 Sep 29;12(10):2801. doi: 10.3390/cancers12102801. Cancers (Basel). 2020. PMID: 33003483 Free PMC article. Review.
References
-
- Tannock, I. F. , and Hickman J. A.. 2016. Limits to personalized cancer medicine. N. Engl. J. Med. 375:1289–1294. - PubMed
-
- Smalley, K. S. 2010. Understanding melanoma signaling networks as the basis for molecular targeted therapy. J. Invest. Dermatol. 130:28–37. - PubMed
-
- Sun, C. , Wang L., Huang S., Heynen G. J. J. E., Prahallad A., Robert C., et al. 2014. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 508:118–122. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

