Plasma metabolome and skin proteins in Charcot-Marie-Tooth 1A patients
- PMID: 28575008
- PMCID: PMC5456076
- DOI: 10.1371/journal.pone.0178376
Plasma metabolome and skin proteins in Charcot-Marie-Tooth 1A patients
Abstract
Objective: Charcot-Marie-Tooth 1A (CMT1A) disease is the most common inherited neuropathy that lacks of therapy and of molecular markers to assess disease severity. Herein, we have pursued the identification of potential biomarkers in plasma samples and skin biopsies that could define the phenotype of CMT1A patients at mild (Mi), moderate (Mo) and severe (Se) stages of disease as assessed by the CMT neuropathy score to contribute to the understanding of CMT pathophysiology and eventually inform of the severity of the disease.
Methods: We have used: (i) a high-throughput untargeted metabolomic approach of plasma samples in a cohort of 42 CMT1A patients and 15 healthy controls (CRL) using ultrahigh liquid chromatography coupled to mass spectrometry and (ii) reverse phase protein microarrays to quantitate the expression of some proteins of energy metabolism and of the antioxidant response in skin biopsies of a cohort of 70 CMT1A patients and 13 healthy controls.
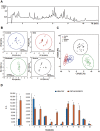
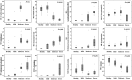
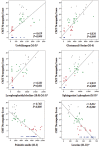
Results: The metabolomic approach identified 194 metabolites with significant differences among the four groups (Mi, Mo, Se, CRL) of samples. A multivariate Linear Discriminant Analysis model using 12 metabolites afforded the correct classification of the samples. These metabolites indicate an increase in protein catabolism and the mobilization of membrane lipids involved in signaling inflammation with severity of CMT1A. A concurrent depletion of leucine, which is required for the biogenesis of the muscle, is also observed in the patients. Protein expression in skin biopsies indicates early loss of mitochondrial and antioxidant proteins in patients' biopsies.
Conclusion: The findings indicate that CMT1A disease is associated with a metabolic state resembling inflammation and sarcopenia suggesting that it might represent a potential target to prevent the nerve and muscle wasting phenotype in these patients. The observed changes in metabolites could be useful as potential biomarkers of CMT1A disease after appropriate validation in future longitudinal studies.
Conflict of interest statement
Figures



References
-
- Lupski JR, de Oca-Luna RM, Slaugenhaupt S, Pentao L, Guzzetta V, Trask BJ, et al. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991;66(2):219–232. . - PubMed
-
- Raeymaekers P, Timmerman V, Nelis E, De Jonghe P, Hoogendijk JE, Baas F, et al. Duplication in chromosome 17p11.2 in Charcot-Marie-Tooth neuropathy type 1a (CMT 1a). The HMSN Collaborative Research Group. Neuromuscul Disord. 1991;1(2):93–97. . - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

