Whole-Genome Cardiac DNA Methylation Fingerprint and Gene Expression Analysis Provide New Insights in the Pathogenesis of Chronic Chagas Disease Cardiomyopathy
- PMID: 28575239
- PMCID: PMC5849099
- DOI: 10.1093/cid/cix506
Whole-Genome Cardiac DNA Methylation Fingerprint and Gene Expression Analysis Provide New Insights in the Pathogenesis of Chronic Chagas Disease Cardiomyopathy
Abstract
Background: Chagas disease, caused by the protozoan Trypanosoma cruzi, is endemic in Latin America and affects 10 million people worldwide. Approximately 12000 deaths attributable to Chagas disease occur annually due to chronic Chagas disease cardiomyopathy (CCC), an inflammatory cardiomyopathy presenting with heart failure and arrythmia; 30% of infected subjects develop CCC years after infection. Genetic mechanisms play a role in differential progression to CCC, but little is known about the role of epigenetic modifications in pathological gene expression patterns in CCC patients' myocardium. DNA methylation is the most common modification in the mammalian genome.
Methods: We investigated the impact of genome-wide cardiac DNA methylation on global gene expression in myocardial samples from end-stage CCC patients, compared to control samples from organ donors.
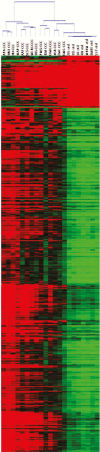
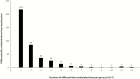
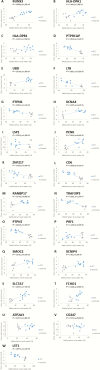
Results: In total, 4720 genes were differentially methylated between CCC patients and controls, of which 399 were also differentially expressed. Several of them were related to heart function or to the immune response and had methylation sites in their promoter region. Reporter gene and in silico transcription factor binding analyses indicated promoter methylation modified expression of key genes. Among those, we found potassium channel genes KCNA4 and KCNIP4, involved in electrical conduction and arrythmia, SMOC2, involved in matrix remodeling, as well as enkephalin and RUNX3, potentially involved in the increased T-helper 1 cytokine-mediated inflammatory damage in heart.
Conclusions: Results support that DNA methylation plays a role in the regulation of expression of pathogenically relevant genes in CCC myocardium, and identify novel potential disease pathways and therapeutic targets in CCC.
Figures


References
-
- Nunes MC, Dones W, Morillo CA, Encina JJ, Ribeiro AL; Council on Chagas Disease of the Interamerican Society of Cardiology Chagas disease: an overview of clinical and epidemiological aspects. J Am Coll Cardiol 2013; 62:767–76. - PubMed
-
- Abel LC, Rizzo LV, Ianni B et al. Chronic Chagas’ disease cardiomyopathy patients display an increased IFN-gamma response to Trypanosoma cruzi infection. J Autoimmun 2001; 17:99–107. - PubMed
-
- Cunha-Neto E, Nogueira LG, Teixeira PC et al. Immunological and non-immunological effects of cytokines and chemokines in the pathogenesis of chronic Chagas disease cardiomyopathy. Mem Inst Oswaldo Cruz 2009; 104(suppl 1):252–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

