Selective disruption of an oncogenic mutant allele by CRISPR/Cas9 induces efficient tumor regression
- PMID: 28575452
- PMCID: PMC5570104
- DOI: 10.1093/nar/gkx490
Selective disruption of an oncogenic mutant allele by CRISPR/Cas9 induces efficient tumor regression
Erratum in
-
Correction to 'Selective disruption of an oncogenic mutant allele by CRISPR/Cas9 induces efficient tumor regression'.Nucleic Acids Res. 2025 Jul 19;53(14):gkaf768. doi: 10.1093/nar/gkaf768. Nucleic Acids Res. 2025. PMID: 40747684 Free PMC article. No abstract available.
Abstract
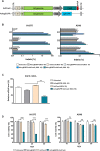
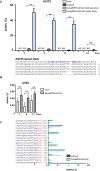
Approximately 15% of non-small cell lung cancer cases are associated with a mutation in the epidermal growth factor receptor (EGFR) gene, which plays a critical role in tumor progression. With the goal of treating mutated EGFR-mediated lung cancer, we demonstrate the use of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9 (Cas9) system to discriminate between the oncogenic mutant and wild-type EGFR alleles and eliminate the carcinogenic mutant EGFR allele with high accuracy. We targeted an EGFR oncogene harboring a single-nucleotide missense mutation (CTG > CGG) that generates a protospacer-adjacent motif sequence recognized by the CRISPR/Cas9 derived from Streptococcus pyogenes. Co-delivery of Cas9 and an EGFR mutation-specific single-guide RNA via adenovirus resulted in precise disruption at the oncogenic mutation site with high specificity. Furthermore, this CRISPR/Cas9-mediated mutant allele disruption led to significantly enhanced cancer cell killing and reduced tumor size in a xenograft mouse model of human lung cancer. Taken together, these results indicate that targeting an oncogenic mutation using CRISPR/Cas9 offers a powerful surgical strategy to disrupt oncogenic mutations to treat cancers; similar strategies could be used to treat other mutation-associated diseases.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Siegel R.L., Miller K.D., Jemal A.. Cancer Statistics, 2017. CA Cancer J. Clin. 2017; 67:7–30. - PubMed
-
- Edwards B.K., Noone A.M., Mariotto A.B., Simard E.P., Boscoe F.P., Henley S.J., Jemal A., Cho H., Anderson R.N., Kohler B.A. et al. Annual Report to the Nation on the status of cancer, 1975–2010, featuring prevalence of comorbidity and impact on survival among persons with lung, colorectal, breast, or prostate cancer. Cancer. 2014; 120:1290–1314. - PMC - PubMed
-
- Ohsaki Y., Tanno S., Fujita Y., Toyoshima E., Fujiuchi S., Nishigaki Y., Ishida S., Nagase A., Miyokawa N., Hirata S. et al. Epidermal growth factor receptor expression correlates with poor prognosis in non-small cell lung cancer patients with p53 overexpression. Oncol. Rep. 2000; 7:603–607. - PubMed
-
- Inamura K., Ninomiya H., Ishikawa Y., Matsubara O.. Is the epidermal growth factor receptor status in lung cancers reflected in clinicopathologic features. Arch. Pathol. Lab. Med. 2010; 134:66–72. - PubMed
-
- Hynes N.E., Lane H.A.. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer. 2005; 5:341–354. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

