Chloroformate derivatization for tracing the fate of Amino acids in cells and tissues by multiple stable isotope resolved metabolomics (mSIRM)
- PMID: 28576319
- PMCID: PMC5638134
- DOI: 10.1016/j.aca.2017.04.014
Chloroformate derivatization for tracing the fate of Amino acids in cells and tissues by multiple stable isotope resolved metabolomics (mSIRM)
Abstract
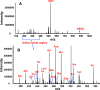
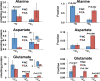
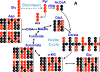
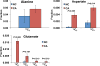
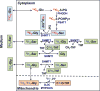
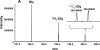
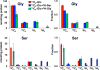
Amino acids have crucial roles in central metabolism, both anabolic and catabolic. To elucidate these roles, steady-state concentrations of amino acids alone are insufficient, as each amino acid participates in multiple pathways and functions in a complex network, which can also be compartmentalized. Stable Isotope-Resolved Metabolomics (SIRM) is an approach that uses atom-resolved tracking of metabolites through biochemical transformations in cells, tissues, or whole organisms. Using different elemental stable isotopes to label multiple metabolite precursors makes it possible to resolve simultaneously the utilization of these precursors in a single experiment. Conversely, a single precursor labeled with two (or more) different elemental isotopes can trace the allocation of e.g. C and N atoms through the network. Such dual-label experiments however challenge the resolution of conventional mass spectrometers, which must distinguish the neutron mass differences among different elemental isotopes. This requires ultrahigh resolution Fourier transform mass spectrometry (UHR-FTMS). When combined with direct infusion nano-electrospray ion source (nano-ESI), UHR-FTMS can provide rapid, global, and quantitative analysis of all possible mass isotopologues of metabolites. Unfortunately, very low mass polar metabolites such as amino acids can be difficult to analyze by current models of UHR-FTMS, plus the high salt content present in typical cell or tissue polar extracts may cause unacceptable ion suppression for sources such as nano-ESI. Here we describe a modified method of ethyl chloroformate (ECF) derivatization of amino acids to enable rapid quantitative analysis of stable isotope labeled amino acids using nano-ESI UHR-FTMS. This method showed excellent linearity with quantifiable limits in the low nanomolar range represented in microgram quantities of biological specimens, which results in extracts with total analyte abundances in the low to sub-femtomole range. We have applied this method to profile amino acids and their labeling patterns in 13C and 2H doubly labeled PC9 cell extracts, cancerous and non-cancerous tissue extracts from a lung cancer patient and their protein hydrolysates as well as plasma extracts from mice fed with a liquid diet containing 13C6-glucose (Glc). The multi-element isotopologue distributions provided key insights into amino acid metabolism and intracellular pools in human lung cancer tissues in high detail. The 13C labeling of Asp and Glu revealed de novo synthesis of these amino acids from 13C6-Glc via the Krebs cycle, specifically the elevated level of 13C3-labeled Asp and Glu in cancerous versus non-cancerous lung tissues was consistent with enhanced pyruvate carboxylation. In addition, tracking the fate of double tracers, (13C6-Glc + 2H2-Gly or 13C6-Glc + 2H3-Ser) in PC9 cells clearly resolved pools of Ser and Gly synthesized de novo from 13C6-Glc (13C3-Ser and 13C2-Gly) versus Ser and Gly derived from external sources (2H3-Ser, 2H2-Gly). Moreover the complex 2H labeling patterns of the latter were results of Ser and Gly exchange through active Ser-Gly one-carbon metabolic pathway in PC9 cells.
Keywords: Amino acids; Direct infusion nano-electrospray; Stable isotope resolved metabolomics; Ultrahigh resolution fourier transform mass spectrometry.
Copyright © 2017 Elsevier B.V. All rights reserved.
Figures



References
-
- Lehninger AL, Nelson DL, Cox MM. Lehninger principles of biochemistry. New York: Worth Publishers; 2000.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

