miR-210 expression is associated with methionine-induced differentiation of trout satellite cells
- PMID: 28576820
- PMCID: PMC6514451
- DOI: 10.1242/jeb.154484
miR-210 expression is associated with methionine-induced differentiation of trout satellite cells
Abstract
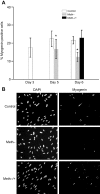
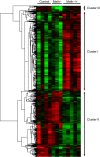
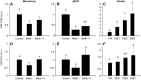
In fish, data on microRNAs (miRNAs) involved in myogenesis are scarce. In order to identify miRNAs involved in satellite cell differentiation, we used a methionine depletion/replenishment protocol to synchronize myogenic cell differentiation. Our results validated that methionine removal (72 h) from the medium strongly decreased myoD1 and myogenin expression, indicating differentiation arrest. In contrast, methionine replenishment rescued expression of myoD1 and myogenin, showing a resumption of differentiation. We performed a miRNA array analysis of myogenic cells under three conditions: presence of methionine for 72 h (control), absence of methionine for 72 h (Meth-) and absence of methionine for 48 h followed by 24 h of methionine replenishment (Meth-/+). A clustering analysis identified three clusters: cluster I corresponds to miRNA upregulated only in Meth-/+ conditions; cluster II corresponds to miRNA downregulated only in Meth-/+ conditions; cluster III corresponds to miRNAs with high expression in control, low expression in Meth- conditions and intermediate expression after methionine replenishment (Meth-/+). Cluster III was very interesting because it fitted with the data obtained for myoD1 and myogenin (supporting an involvement in differentiation) and contained seven miRNAs with muscle-related function (e.g. miR-133a) and one (miR-210) with unknown function. Based on our previously published miRNA repertoire ( Juanchich et al., 2016), we confirmed miR-133a was expressed only in white muscle and showed that miR-210 had strong expression in white muscle. We also showed that miR-210 expression was upregulated during differentiation of satellite cells, suggesting that miR-210 was potentially involved in the differentiation of satellite cells.
Keywords: Methionine depletion; Myo-miR; Myoblast; Myogenesis; miRNA.
© 2017. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

