The Role of CtBP1 in Oncogenic Processes and Its Potential as a Therapeutic Target
- PMID: 28576945
- PMCID: PMC5458631
- DOI: 10.1158/1535-7163.MCT-16-0592
The Role of CtBP1 in Oncogenic Processes and Its Potential as a Therapeutic Target
Abstract
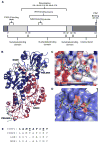
Transcriptional corepressor proteins have emerged as an important facet of cancer etiology. These corepressor proteins are often altered by loss- or gain-of-function mutations, leading to transcriptional imbalance. Thus, research directed at expanding our current understanding of transcriptional corepressors could impact the future development of new cancer diagnostics, prognostics, and therapies. In this review, our current understanding of the CtBP corepressors, and their role in both development and disease, is discussed in detail. Importantly, the role of CtBP1 overexpression in adult tissues in promoting the progression of multiple cancer types through their ability to modulate the transcription of developmental genes ectopically is explored. CtBP1 overexpression is known to be protumorigenic and affects the regulation of gene networks associated with "cancer hallmarks" and malignant behavior, including increased cell survival, proliferation, migration, invasion, and the epithelial-mesenchymal transition. As a transcriptional regulator of broad developmental processes capable of promoting malignant growth in adult tissues, therapeutically targeting the CtBP1 corepressor has the potential to be an effective method for the treatment of diverse tumor types. Although efforts to develop CtBP1 inhibitors are still in the early stages, the current progress and the future perspectives of therapeutically targeting this transcriptional corepressor are also discussed. Mol Cancer Ther; 16(6); 981-90. ©2017 AACR.
©2017 American Association for Cancer Research.
Conflict of interest statement
No potential conflicts of interest.
Figures
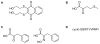
Similar articles
-
The transcriptional corepressor CtBP: a foe of multiple tumor suppressors.Cancer Res. 2009 Feb 1;69(3):731-4. doi: 10.1158/0008-5472.CAN-08-3349. Epub 2009 Jan 20. Cancer Res. 2009. PMID: 19155295 Free PMC article. Review.
-
CtBP- an emerging oncogene and novel small molecule drug target: Advances in the understanding of its oncogenic action and identification of therapeutic inhibitors.Cancer Biol Ther. 2017 Jun 3;18(6):379-391. doi: 10.1080/15384047.2017.1323586. Epub 2017 May 22. Cancer Biol Ther. 2017. PMID: 28532298 Free PMC article. Review.
-
Identification and characterization of a new potent inhibitor targeting CtBP1/BARS in melanoma cells.J Exp Clin Cancer Res. 2024 May 6;43(1):137. doi: 10.1186/s13046-024-03044-5. J Exp Clin Cancer Res. 2024. PMID: 38711119 Free PMC article.
-
Active-Site Tryptophan, the Target of Antineoplastic C-Terminal Binding Protein Inhibitors, Mediates Inhibitor Disruption of CtBP Oligomerization and Transcription Coregulatory Activities.Mol Pharmacol. 2019 Jul;96(1):99-108. doi: 10.1124/mol.118.114363. Epub 2019 Apr 29. Mol Pharmacol. 2019. PMID: 31036695 Free PMC article.
-
Role of transcriptional corepressor CtBP1 in prostate cancer progression.Neoplasia. 2012 Oct;14(10):905-14. doi: 10.1593/neo.121192. Neoplasia. 2012. PMID: 23097625 Free PMC article.
Cited by
-
Transcriptional corepressor activity of CtBP1 is regulated by ISG15 modification.Anim Cells Syst (Seoul). 2024 Feb 22;28(1):66-74. doi: 10.1080/19768354.2024.2321354. eCollection 2024. Anim Cells Syst (Seoul). 2024. PMID: 38405356 Free PMC article.
-
C-Terminal Binding Protein 1 Modulates Cellular Redox via Feedback Regulation of MPC1 and MPC2 in Melanoma Cells.Med Sci Monit. 2018 Oct 25;24:7614-7624. doi: 10.12659/MSM.912735. Med Sci Monit. 2018. PMID: 30356033 Free PMC article.
-
CTBP1 depletion on prostate tumors deregulates miRNA/mRNA expression and impairs cancer progression in metabolic syndrome mice.Cell Death Dis. 2019 Apr 1;10(4):299. doi: 10.1038/s41419-019-1535-z. Cell Death Dis. 2019. PMID: 30931931 Free PMC article.
-
CtBP1 interacts with SOX2 to promote the growth, migration and invasion of lung adenocarcinoma.Oncol Rep. 2019 Jul;42(1):67-78. doi: 10.3892/or.2019.7142. Epub 2019 May 2. Oncol Rep. 2019. Retraction in: Oncol Rep. 2023 Aug;50(2):151. doi: 10.3892/or.2023.8588. PMID: 31059077 Free PMC article. Retracted.
-
Mechanisms contributing to persistently activated cell phenotypes in pulmonary hypertension.J Physiol. 2019 Feb;597(4):1103-1119. doi: 10.1113/JP275857. Epub 2018 Aug 7. J Physiol. 2019. PMID: 29920674 Free PMC article. Review.
References
-
- Chinnadurai G. Transcriptional regulation by C-terminal binding proteins. Int J Biochem Cell Biol. 2007;39(9):1593–607. - PubMed
-
- Spano S, Silletta MG, Colanzi A, Alberti S, Fiucci G, Valente C, et al. Molecular cloning and functional characterization of brefeldin A-ADP-ribosylated substrate. A novel protein involved in the maintenance of the Golgi structure. J Biol Chem. 1999 Jun 18;274(25):17705–10. - PubMed
-
- Katsanis N, Fisher EM. A novel C-terminal binding protein (CTBP2) is closely related to CTBP1, an adenovirus E1A-binding protein, and maps to human chromosome 21q21.3. Genomics. 1998 Jan 15;47(2):294–9. - PubMed
-
- Schmitz F, Konigstorfer A, Sudhof TC. RIBEYE, a component of synaptic ribbons: a protein’s journey through evolution provides insight into synaptic ribbon function. Neuron. 2000 Dec;28(3):857–72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

