Glycosylation and oligomeric state of envelope protein might influence HIV-1 virion capture by α4β7 integrin
- PMID: 28577856
- PMCID: PMC5526109
- DOI: 10.1016/j.virol.2017.05.016
Glycosylation and oligomeric state of envelope protein might influence HIV-1 virion capture by α4β7 integrin
Abstract
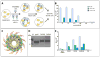
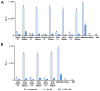
The α4ß7 integrin present on host cells recognizes the V1V2 domain of the HIV-1 envelope protein. This interaction might be involved in virus transmission. Administration of α4ß7-specific antibodies inhibit acquisition of SIV in a macaque challenge model. But the molecular details of V1V2: α4ß7 interaction are unknown and its importance in HIV-1 infection remains controversial. Our biochemical and mutational analyses show that glycosylation is a key modulator of V1V2 conformation and binding to α4ß7. Partially glycosylated, but not fully glycosylated, envelope proteins are preferred substrates for α4ß7 binding. Surprisingly, monomers of the envelope protein bound strongly to α4ß7 whereas trimers bound poorly. Our results suggest that a conformationally flexible V1V2 domain allows binding of the HIV-1 virion to the α4ß7 integrin, which might impart selectivity for the poorly glycosylated HIV-1 envelope containing monomers to be more efficiently captured by α4ß7 integrin present on mucosal cells at the time of HIV-1 transmission.
Keywords: Envelope glycoprotein; HIV vaccine; HIV-1; V1V2 domain; Virus capture; Virus entry; α4β7 integrin.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures




Similar articles
-
Envelope glycoprotein binding to the integrin α4β7 is not a general property of most HIV-1 strains.J Virol. 2014 Sep;88(18):10767-77. doi: 10.1128/JVI.03296-13. Epub 2014 Jul 9. J Virol. 2014. PMID: 25008916 Free PMC article.
-
Common helical V1V2 conformations of HIV-1 Envelope expose the α4β7 binding site on intact virions.Nat Commun. 2018 Oct 26;9(1):4489. doi: 10.1038/s41467-018-06794-x. Nat Commun. 2018. PMID: 30367034 Free PMC article.
-
The V1V2 Region of HIV-1 gp120 Forms a Five-Stranded Beta Barrel.J Virol. 2015 Aug;89(15):8003-10. doi: 10.1128/JVI.00754-15. Epub 2015 May 27. J Virol. 2015. PMID: 26018158 Free PMC article.
-
HIV-1 envelope, integrins and co-receptor use in mucosal transmission of HIV.J Transl Med. 2011 Jan 27;9 Suppl 1(Suppl 1):S2. doi: 10.1186/1479-5876-9-S1-S2. J Transl Med. 2011. PMID: 21284901 Free PMC article. Review.
-
Integrin α4β7 in HIV-1 infection: A critical review.J Leukoc Biol. 2020 Aug;108(2):627-632. doi: 10.1002/JLB.4MR0120-208R. Epub 2020 Apr 9. J Leukoc Biol. 2020. PMID: 32272507 Review.
Cited by
-
Abiological catalysis by myoglobin mutant with a genetically incorporated unnatural amino acid.Biochem J. 2021 May 14;478(9):1795-1808. doi: 10.1042/BCJ20210091. Biochem J. 2021. PMID: 33821889 Free PMC article.
-
V2-Specific Antibodies in HIV-1 Vaccine Research and Natural Infection: Controllers or Surrogate Markers.Vaccines (Basel). 2019 Aug 6;7(3):82. doi: 10.3390/vaccines7030082. Vaccines (Basel). 2019. PMID: 31390725 Free PMC article. Review.
-
The V2 domain of HIV gp120 mimics an interaction between CD4 and integrin ⍺4β7.PLoS Pathog. 2023 Dec 8;19(12):e1011860. doi: 10.1371/journal.ppat.1011860. eCollection 2023 Dec. PLoS Pathog. 2023. PMID: 38064524 Free PMC article.
-
Methamphetamine Induces the Release of Proadhesive Extracellular Vesicles and Promotes Syncytia Formation: A Potential Role in HIV-1 Neuropathogenesis.Viruses. 2022 Mar 7;14(3):550. doi: 10.3390/v14030550. Viruses. 2022. PMID: 35336957 Free PMC article.
-
Multimeric Epitope-Scaffold HIV Vaccines Target V1V2 and Differentially Tune Polyfunctional Antibody Responses.Cell Rep. 2019 Jul 23;28(4):877-895.e6. doi: 10.1016/j.celrep.2019.06.074. Cell Rep. 2019. PMID: 31340151 Free PMC article.
References
-
- Ansari AA, Reimann KA, Mayne AE, Takahashi Y, Stephenson ST, Wang R, … Villinger F. Blocking of alpha4beta7 gut-homing integrin during acute infection leads to decreased plasma and gastrointestinal tissue viral loads in simian immunodeficiency virus-infected rhesus macaques. J Immunol. 2011;186(2):1044–1059. doi: 10.4049/jimmunol.1003052. - DOI - PMC - PubMed
-
- Beddows S, Schulke N, Kirschner M, Barnes K, Franti M, Michael E, … Moore JP. Evaluating the immunogenicity of a disulfide-stabilized, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2005;79(14):8812–8827. doi: 10.1128/JVI.79.14.8812-8827.2005. - DOI - PMC - PubMed
-
- Berlin C, Berg EL, Briskin MJ, Andrew DP, Kilshaw PJ, Holzmann B, … Butcher EC. Alpha 4 beta 7 integrin mediates lymphocyte binding to the mucosal vascular addressin MAdCAM-1. Cell. 1993;74(1):185–195. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

