De-identification of clinical notes via recurrent neural network and conditional random field
- PMID: 28579533
- PMCID: PMC5705329
- DOI: 10.1016/j.jbi.2017.05.023
De-identification of clinical notes via recurrent neural network and conditional random field
Abstract
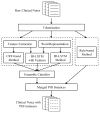
De-identification, identifying information from data, such as protected health information (PHI) present in clinical data, is a critical step to enable data to be shared or published. The 2016 Centers of Excellence in Genomic Science (CEGS) Neuropsychiatric Genome-scale and RDOC Individualized Domains (N-GRID) clinical natural language processing (NLP) challenge contains a de-identification track in de-identifying electronic medical records (EMRs) (i.e., track 1). The challenge organizers provide 1000 annotated mental health records for this track, 600 out of which are used as a training set and 400 as a test set. We develop a hybrid system for the de-identification task on the training set. Firstly, four individual subsystems, that is, a subsystem based on bidirectional LSTM (long-short term memory, a variant of recurrent neural network), a subsystem-based on bidirectional LSTM with features, a subsystem based on conditional random field (CRF) and a rule-based subsystem, are used to identify PHI instances. Then, an ensemble learning-based classifiers is deployed to combine all PHI instances predicted by above three machine learning-based subsystems. Finally, the results of the ensemble learning-based classifier and the rule-based subsystem are merged together. Experiments conducted on the official test set show that our system achieves the highest micro F1-scores of 93.07%, 91.43% and 95.23% under the "token", "strict" and "binary token" criteria respectively, ranking first in the 2016 CEGS N-GRID NLP challenge. In addition, on the dataset of 2014 i2b2 NLP challenge, our system achieves the highest micro F1-scores of 96.98%, 95.11% and 98.28% under the "token", "strict" and "binary token" criteria respectively, outperforming other state-of-the-art systems. All these experiments prove the effectiveness of our proposed method.
Keywords: De-identification; Ensemble system; Natural language processing; Protected health information; Recurrent neural network.
Copyright © 2017. Published by Elsevier Inc.
Figures


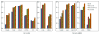
References
-
- A. Act. Health insurance portability and accountability act of 1996. Public Law. 1996;104:191. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

