Exploiting a host-commensal interaction to promote intestinal barrier function and enteric pathogen tolerance
- PMID: 28580440
- PMCID: PMC5453653
- DOI: 10.1126/sciimmunol.aai7732
Exploiting a host-commensal interaction to promote intestinal barrier function and enteric pathogen tolerance
Abstract
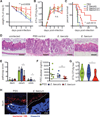
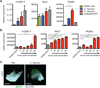
Commensal intestinal bacteria can prevent pathogenic infection; however, limited knowledge of the mechanisms by which individual bacterial species contribute to pathogen resistance has restricted their potential for therapeutic application. Here, we examined how colonization of mice with a human commensal Enterococcus faecium protects against enteric infections. We show that E. faecium improves host intestinal epithelial defense programs to limit Salmonella enterica serotype Typhimurium pathogenesis in vivo in multiple models of susceptibility. E. faecium protection is mediated by a unique peptidoglycan hydrolase, SagA, and requires epithelial expression of pattern recognition receptor components and antimicrobial peptides. Ectopic expression of SagA in non-protective and probiotic bacteria is sufficient to enhance intestinal barrier function and confer resistance against S. Typhimurium and Clostridium difficile pathogenesis. These studies demonstrate that specific factors from commensal bacteria can be used to improve host barrier function and limit the pathogenesis of distinct enteric infections.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

