The ribosome and its role in protein folding: looking through a magnifying glass
- PMID: 28580913
- PMCID: PMC5458493
- DOI: 10.1107/S2059798317007446
The ribosome and its role in protein folding: looking through a magnifying glass
Abstract
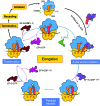
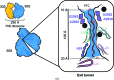
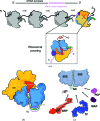
Protein folding, a process that underpins cellular activity, begins co-translationally on the ribosome. During translation, a newly synthesized polypeptide chain enters the ribosomal exit tunnel and actively interacts with the ribosome elements - the r-proteins and rRNA that line the tunnel - prior to emerging into the cellular milieu. While understanding of the structure and function of the ribosome has advanced significantly, little is known about the process of folding of the emerging nascent chain (NC). Advances in cryo-electron microscopy are enabling visualization of NCs within the exit tunnel, allowing early glimpses of the interplay between the NC and the ribosome. Once it has emerged from the exit tunnel into the cytosol, the NC (still attached to its parent ribosome) can acquire a range of conformations, which can be characterized by NMR spectroscopy. Using experimental restraints within molecular-dynamics simulations, the ensemble of NC structures can be described. In order to delineate the process of co-translational protein folding, a hybrid structural biology approach is foreseeable, potentially offering a complete atomic description of protein folding as it occurs on the ribosome.
Keywords: NMR; cryo-EM; nascent chain; protein folding; ribosome.
Figures



Similar articles
-
alpha-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel.Nat Struct Mol Biol. 2010 Mar;17(3):313-7. doi: 10.1038/nsmb.1756. Epub 2010 Feb 7. Nat Struct Mol Biol. 2010. PMID: 20139981
-
Cryo-EM Structures Reveal Relocalization of MetAP in the Presence of Other Protein Biogenesis Factors at the Ribosomal Tunnel Exit.J Mol Biol. 2019 Mar 29;431(7):1426-1439. doi: 10.1016/j.jmb.2019.02.002. Epub 2019 Feb 10. J Mol Biol. 2019. PMID: 30753870
-
How Does the Ribosome Fold the Proteome?Annu Rev Biochem. 2020 Jun 20;89:389-415. doi: 10.1146/annurev-biochem-062917-012226. Annu Rev Biochem. 2020. PMID: 32569518 Review.
-
Protein folding on the ribosome studied using NMR spectroscopy.Prog Nucl Magn Reson Spectrosc. 2013 Oct;74(100):57-75. doi: 10.1016/j.pnmrs.2013.07.003. Epub 2013 Jul 27. Prog Nucl Magn Reson Spectrosc. 2013. PMID: 24083462 Free PMC article. Review.
-
A structural ensemble of a ribosome-nascent chain complex during cotranslational protein folding.Nat Struct Mol Biol. 2016 Apr;23(4):278-285. doi: 10.1038/nsmb.3182. Epub 2016 Feb 29. Nat Struct Mol Biol. 2016. PMID: 26926436 Free PMC article.
Cited by
-
Unraveling co-translational protein folding: Concepts and methods.Methods. 2018 Mar 15;137:71-81. doi: 10.1016/j.ymeth.2017.11.007. Epub 2017 Dec 6. Methods. 2018. PMID: 29221924 Free PMC article. Review.
-
Conserved residues Glu37 and Trp229 play an essential role in protein folding of β-lactamase.FEBS J. 2021 Oct;288(19):5708-5722. doi: 10.1111/febs.15854. Epub 2021 May 2. FEBS J. 2021. PMID: 33792206 Free PMC article.
-
Integration of text mining and biological network analysis: Identification of essential genes in sulfate-reducing bacteria.Front Microbiol. 2023 Apr 13;14:1086021. doi: 10.3389/fmicb.2023.1086021. eCollection 2023. Front Microbiol. 2023. PMID: 37125195 Free PMC article.
-
Interactome Mapping of eIF3A in a Colon Cancer and an Immortalized Embryonic Cell Line Using Proximity-Dependent Biotin Identification.Cancers (Basel). 2021 Mar 14;13(6):1293. doi: 10.3390/cancers13061293. Cancers (Basel). 2021. PMID: 33799492 Free PMC article.
-
Differences in the path to exit the ribosome across the three domains of life.Nucleic Acids Res. 2019 May 7;47(8):4198-4210. doi: 10.1093/nar/gkz106. Nucleic Acids Res. 2019. PMID: 30805621 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

