Dystrophin-glycoprotein complex sequesters Yap to inhibit cardiomyocyte proliferation
- PMID: 28581498
- PMCID: PMC5528853
- DOI: 10.1038/nature22979
Dystrophin-glycoprotein complex sequesters Yap to inhibit cardiomyocyte proliferation
Abstract
The regenerative capacity of the adult mammalian heart is limited, because of the reduced ability of cardiomyocytes to progress through mitosis. Endogenous cardiomyocytes have regenerative capacity at birth but this capacity is lost postnatally, with subsequent organ growth occurring through cardiomyocyte hypertrophy. The Hippo pathway, a conserved kinase cascade, inhibits cardiomyocyte proliferation in the developing heart to control heart size and prevents regeneration in the adult heart. The dystrophin-glycoprotein complex (DGC), a multicomponent transmembrane complex linking the actin cytoskeleton to extracellular matrix, is essential for cardiomyocyte homeostasis. DGC deficiency in humans results in muscular dystrophy, including the lethal Duchenne muscular dystrophy. Here we show that the DGC component dystroglycan 1 (Dag1) directly binds to the Hippo pathway effector Yap to inhibit cardiomyocyte proliferation in mice. The Yap-Dag1 interaction was enhanced by Hippo-induced Yap phosphorylation, revealing a connection between Hippo pathway function and the DGC. After injury, Hippo-deficient postnatal mouse hearts maintained organ size control by repairing the defect with correct dimensions, whereas postnatal hearts deficient in both Hippo and the DGC showed cardiomyocyte overproliferation at the injury site. In the hearts of mature Mdx mice (which have a point mutation in Dmd)-a model of Duchenne muscular dystrophy-Hippo deficiency protected against overload-induced heart failure.
Figures
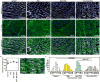



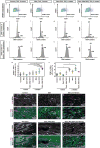
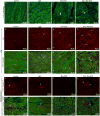

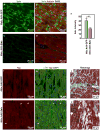

Hippo signaling is low. YAP phosphorylation and YAP binding to the DGC is reduced.
YAP-TEAD promote the transcription of target genes including Sgcδ and α-catenin.
YAP-TEAD promote DGC assembly by promoting the expression of the core component Sgcδ.
The ICD is immature in neonatal cardiomyocytes. Yap promotes the expression of the ICD component α3-catenin.
Hippo signaling is high. YAP phosphorylation and YAP binding to the DGC is increased.
YAP-TEAD transcription-activating activity is reduced.
The DGC sequesters phosphorylated YAP through an interaction involving the PPxY motif of DAG1.
The ICD is mature in adult cardiomyocytes. YAP is incorporated into the ICD independent of Hippo through α-catenin binding.




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

