Mechanisms of suppression: The wiring of genetic resilience
- PMID: 28582599
- PMCID: PMC5681848
- DOI: 10.1002/bies.201700042
Mechanisms of suppression: The wiring of genetic resilience
Abstract
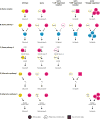
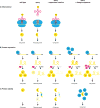
Recent analysis of genome sequences has identified individuals that are healthy despite carrying severe disease-associated mutations. A possible explanation is that these individuals carry a second genomic perturbation that can compensate for the detrimental effects of the disease allele, a phenomenon referred to as suppression. In model organisms, suppression interactions are generally divided into two classes: genomic suppressors which are secondary mutations in the genome that bypass a mutant phenotype, and dosage suppression interactions in which overexpression of a suppressor gene rescues a mutant phenotype. Here, we describe the general properties of genomic and dosage suppression, with an emphasis on the budding yeast. We propose that suppression interactions between genetic variants are likely relevant for determining the penetrance of human traits. Consequently, an understanding of suppression mechanisms may guide the discovery of protective variants in healthy individuals that carry disease alleles, which could direct the rational design of new therapeutics.
Keywords: compensatory evolution; dosage suppression; epistasis; genetic interactions; protective alleles; suppression interactions; synthetic viability.
© 2017 WILEY Periodicals, Inc.
Figures

References
-
- Nadeau JH. Modifier genes in mice and humans. Nat Rev Genet. 2001;2:165–74. - PubMed
-
- Harper AR, Nayee S, Topol EJ. Protective alleles and modifier variants in human health and disease. Nat Rev Genet. 2015;16:689–701. - PubMed
-
- Chen R, Shi L, Hakenberg J, Naughton B, et al. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat Biotechnol. 2016;34:531–8. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

