Prevalence and distribution of Gardnerella vaginalis subgroups in women with and without bacterial vaginosis
- PMID: 28583109
- PMCID: PMC5460423
- DOI: 10.1186/s12879-017-2501-y
Prevalence and distribution of Gardnerella vaginalis subgroups in women with and without bacterial vaginosis
Abstract
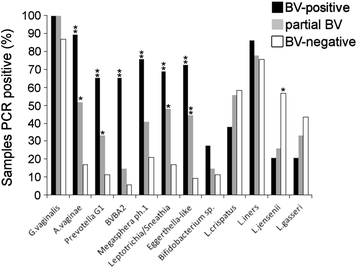
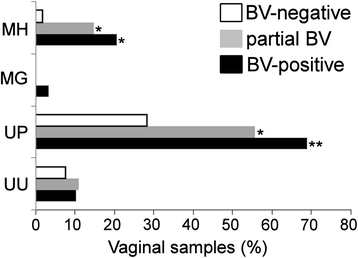
Background: Bacterial vaginosis (BV) is one of the leading causes of vaginal complaints among women of childbearing age. The role of Gardnerella vaginalis remains controversial due to its presence in healthy and BV-type vaginal microflora. The phenotypic and genotypic heterogeneity of G. vaginalis suggested the existence of strain variants linked with different health conditions. We sought to analyze prevalence and distribution of G. vaginalis subgroups (clades) in BV-positive (n = 29), partial BV (n = 27), and BV-negative (n = 53) vaginal samples from Lithuanian women.
Methods: Vaginal samples were characterized by Amsel criteria and the Nugent method. Bacterial signatures characteristic of BV and concomitant infections were identified by culture and PCR. Using singleplex PCR assays, G. vaginalis subgroups were identified in 109 noncultured vaginal specimens by targeting clade-specific genes. Isolated G. vaginalis clinical strains were subtyped and the presence of the sialidase coding gene was detected by PCR. Data analysis was performed using GraphPad Prism statistical software.
Results: G. vaginalis was found in 87% of women without BV. Clade 4 was most frequently detected (79.4%), followed by clade 1 (63.7%), clade 2 (42.2%), and clade 3 (15.7%). Multi-clade G. vaginalis communities showed a positive association with Nugent score (NS) ≥ 4 (OR 3.64; 95% CI 1.48-8.91; p = 0.005). Clade 1 and clade 2 were statistically significantly more common in samples with NS 7-10 (OR 4.69; 95% CI 1.38-15.88; p = 0.01 and OR 6.26; 95% CI 2.20-17.81; p ≤ 0.001, respectively). Clade 3 and clade 4 showed no association with high NS (OR 0.88; 95% CI 0.26-3.04; p = 1.00 and OR 1.31; 95% CI 0.39-4.41; p = 0.767, respectively). The gene coding for sialidase was detected in all isolates of clade 1 and clade 2, but not in clade 4 isolates.
Conclusions: We showed an association between the microbial state of vaginal microflora and specific subgroups of G. vaginalis, the distribution of which may determine the clinical manifestation of BV. The frequent detection of clade 4 in the BV-negative samples might be due its lack of the gene coding for sialidase.
Keywords: Bacterial vaginosis; Clade; Gardnerella vaginalis; Sialidase A; Subgroup; Vaginal microbiota.
Figures

References
-
- Srinivasan S, Hoffman NG, Morgan MT, Matsen FA, Fiedler TL, Hall RW, et al. Bacterial communities in women with bacterial vaginosis: high resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PLoS One. 2012;7(6) doi: 10.1371/journal.pone.0037818. - DOI - PMC - PubMed
-
- Bilardi JE, Walker S, Temple-Smith M, McNair R, Mooney-Somers J, Bellhouse C, et al. The burden of bacterial vaginosis: women‘s experience of the physical, emotional, sexual and social impact of living with recurrent bacterial vaginosis. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0074378. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

