Fine mapping by composite genome-wide association analysis
- PMID: 28583209
- PMCID: PMC6865146
- DOI: 10.1017/S0016672317000027
Fine mapping by composite genome-wide association analysis
Abstract
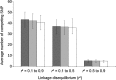
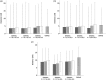
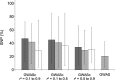
Genome-wide association (GWA) studies play a key role in current genetics research, unravelling genomic regions linked to phenotypic traits of interest in multiple species. Nevertheless, the extent of linkage disequilibrium (LD) may provide confounding results when significant genetic markers span along several contiguous cM. In this study, we have adapted the composite interval mapping approach to the GWA framework (composite GWA), in order to evaluate the impact of including competing (possibly linked) genetic markers when testing for the additive allelic effect inherent to a given genetic marker. We tested model performance on simulated data sets under different scenarios (i.e., qualitative trait loci effects, LD between genetic markers and width of the genomic region involved in the analysis). Our results showed that the genomic region had a small impact on the number of competing single nucleotide polymorphisms (SNPs) as well as on the precision of the composite GWA analysis. A similar conclusion was derived from the preferable range of LD between the tested SNP and competing SNPs, although moderate-to-high LD seemed to attenuate the loss of statistical power. The composite GWA improved specificity and reduced the number of significant genetic markers. The composite GWA model contributes a novel point of view for GWA analyses where testing circumscribed to the genomic region flanking each SNP (delimited by the nearest competing SNPs) and conditioning on linked markers increases the precision to locate causal mutations, but possibly at the expense of power.
Figures


References
-
- Benjamini Y. & Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B 57, 289–290.
-
- Benyamin B., Visscher P. M. & McRae A. F. (2009). Family-based genome-wide association studies. Pharmacogenomics 10, 181–190. - PubMed
-
- Bernardo R. (2013). Genomewide markers as cofactors for precision mapping of quantitative trait loci. Theoretical and Applied Genetics 126, 999–1009. - PubMed
-
- Bonferroni C. E. (1936). Teoria statistica della classi a calcolo della probabilità. Pubblicasioni del R Instituto Superiore di Scienze Economiche e Commerciali di Firenze 8, 3–62.
-
- Casellas J. & Varona L. (2011). Effect of mutation age on genomic prediction. Journal of Dairy Science 94, 4224–4229. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
