Mutations of PKA cyclic nucleotide-binding domains reveal novel aspects of cyclic nucleotide selectivity
- PMID: 28583991
- PMCID: PMC5896744
- DOI: 10.1042/BCJ20160969
Mutations of PKA cyclic nucleotide-binding domains reveal novel aspects of cyclic nucleotide selectivity
Abstract
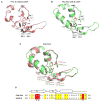
Cyclic AMP and cyclic GMP are ubiquitous second messengers that regulate the activity of effector proteins in all forms of life. The main effector proteins, the 3',5'-cyclic adenosine monophosphate (cAMP)-dependent protein kinase (PKA) and the 3',5'-cyclic guanosine monophosphate (cGMP)-dependent protein kinase (PKG), are preferentially activated by cAMP and cGMP, respectively. However, the molecular basis of this cyclic nucleotide selectivity is still not fully understood. Analysis of isolated cyclic nucleotide-binding (CNB) domains of PKA regulatory subunit type Iα (RIα) reveals that the C-terminal CNB-B has a higher cAMP affinity and selectivity than the N-terminal CNB-A. Here, we show that introducing cGMP-specific residues using site-directed mutagenesis reduces the selectivity of CNB-B, while the combination of two mutations (G316R/A336T) results in a cGMP-selective binding domain. Furthermore, introducing the corresponding mutations (T192R/A212T) into the PKA RIα CNB-A turns this domain into a highly cGMP-selective domain, underlining the importance of these contacts for achieving cGMP specificity. Binding data with the generic purine nucleotide 3',5'-cyclic inosine monophosphate (cIMP) reveal that introduced arginine residues interact with the position 6 oxygen of the nucleobase. Co-crystal structures of an isolated CNB-B G316R/A336T double mutant with either cAMP or cGMP reveal that the introduced threonine and arginine residues maintain their conserved contacts as seen in PKG I CNB-B. These results improve our understanding of cyclic nucleotide binding and the molecular basis of cyclic nucleotide specificity.
Keywords: CNB domain; cAMP; cGMP; cyclic nucleotide; protein kinase A; protein kinase G.
© 2017 The Author(s); published by Portland Press Limited on behalf of the Biochemical Society.
Conflict of interest statement
The Authors declare that there are no competing interests associated with the manuscript.
Figures

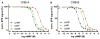




Similar articles
-
Switching Cyclic Nucleotide-Selective Activation of Cyclic Adenosine Monophosphate-Dependent Protein Kinase Holoenzyme Reveals Distinct Roles of Tandem Cyclic Nucleotide-Binding Domains.ACS Chem Biol. 2017 Dec 15;12(12):3057-3066. doi: 10.1021/acschembio.7b00732. Epub 2017 Nov 21. ACS Chem Biol. 2017. PMID: 29111666 Free PMC article.
-
Structural basis for cyclic-nucleotide selectivity and cGMP-selective activation of PKG I.Structure. 2014 Jan 7;22(1):116-24. doi: 10.1016/j.str.2013.09.021. Epub 2013 Nov 14. Structure. 2014. PMID: 24239458 Free PMC article.
-
Contribution of the carboxyl-terminal regional of the cAMP-dependent protein kinase type I alpha regulatory subunit to cyclic nucleotide interactions.Arch Biochem Biophys. 1997 Dec 15;348(2):347-56. doi: 10.1006/abbi.1997.0431. Arch Biochem Biophys. 1997. PMID: 9434747
-
cAMP-Dependent Protein Kinase and cGMP-Dependent Protein Kinase as Cyclic Nucleotide Effectors.Handb Exp Pharmacol. 2017;238:105-122. doi: 10.1007/164_2015_36. Handb Exp Pharmacol. 2017. PMID: 27885524 Review.
-
Cyclic nucleotide selectivity of protein kinase G isozymes.Protein Sci. 2021 Feb;30(2):316-327. doi: 10.1002/pro.4008. Epub 2020 Dec 10. Protein Sci. 2021. PMID: 33271627 Free PMC article. Review.
Cited by
-
The Popeye Domain Containing Genes and Their Function as cAMP Effector Proteins in Striated Muscle.J Cardiovasc Dev Dis. 2018 Mar 13;5(1):18. doi: 10.3390/jcdd5010018. J Cardiovasc Dev Dis. 2018. PMID: 29534001 Free PMC article. Review.
-
Purine nucleosides replace cAMP in allosteric regulation of PKA in trypanosomatid pathogens.Elife. 2024 Mar 22;12:RP91040. doi: 10.7554/eLife.91040. Elife. 2024. PMID: 38517938 Free PMC article.
-
Molecular determinants and signaling effects of PKA RIα phase separation.Mol Cell. 2024 Apr 18;84(8):1570-1584.e7. doi: 10.1016/j.molcel.2024.03.002. Epub 2024 Mar 26. Mol Cell. 2024. PMID: 38537638 Free PMC article.
-
Activation of PKA via asymmetric allosteric coupling of structurally conserved cyclic nucleotide binding domains.Nat Commun. 2019 Sep 4;10(1):3984. doi: 10.1038/s41467-019-11930-2. Nat Commun. 2019. PMID: 31484930 Free PMC article.
-
Combined Multiomics and In Silico Approach Uncovers PRKAR1A as a Putative Therapeutic Target in Multi-Organ Dysfunction Syndrome.ACS Omega. 2023 Mar 1;8(10):9555-9568. doi: 10.1021/acsomega.3c00020. eCollection 2023 Mar 14. ACS Omega. 2023. PMID: 36936296 Free PMC article.
References
-
- Sutherland EW, Rall TW. Fractionation and characterization of a cyclic adenine ribonucleotide formed by tissue particles. J Biol Chem. 1958;232:1077–1091. - PubMed
-
- Hofmann F, Ammendola A, Schlossmann J. Rising behind NO: cGMP-dependent protein kinases. J Cell Sci. 2000;113(Pt 10):1671–1676. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

