Mutations in N-acetylglucosamine (O-GlcNAc) transferase in patients with X-linked intellectual disability
- PMID: 28584052
- PMCID: PMC5535036
- DOI: 10.1074/jbc.M117.790097
Mutations in N-acetylglucosamine (O-GlcNAc) transferase in patients with X-linked intellectual disability
Abstract
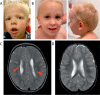
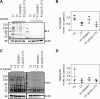
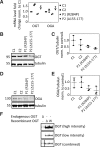
N-Acetylglucosamine (O-GlcNAc) transferase (OGT) regulates protein O-GlcNAcylation, an essential and dynamic post-translational modification. The O-GlcNAc modification is present on numerous nuclear and cytosolic proteins and has been implicated in essential cellular functions such as signaling and gene expression. Accordingly, altered levels of protein O-GlcNAcylation have been associated with developmental defects and neurodegeneration. However, mutations in the OGT gene have not yet been functionally confirmed in humans. Here, we report on two hemizygous mutations in OGT in individuals with X-linked intellectual disability (XLID) and dysmorphic features: one missense mutation (p.Arg284Pro) and one mutation leading to a splicing defect (c.463-6T>G). Both mutations reside in the tetratricopeptide repeats of OGT that are essential for substrate recognition. We observed slightly reduced levels of OGT protein and reduced levels of its opposing enzyme O-GlcNAcase in both patient-derived fibroblasts, but global O-GlcNAc levels appeared to be unaffected. Our data suggest that mutant cells attempt to maintain global O-GlcNAcylation by down-regulating O-GlcNAcase expression. We also found that the c.463-6T>G mutation leads to aberrant mRNA splicing, but no stable truncated protein was detected in the corresponding patient-derived fibroblasts. Recombinant OGT bearing the p.Arg284Pro mutation was prone to unfolding and exhibited reduced glycosylation activity against a complex array of glycosylation substrates and proteolytic processing of the transcription factor host cell factor 1, which is also encoded by an XLID-associated gene. We conclude that defects in O-GlcNAc homeostasis and host cell factor 1 proteolysis may play roles in mediation of XLID in individuals with OGT mutations.
Keywords: Congenital Disorders of Glycosylation; Host Cell Factor 1 (HCF-1); O-GlcNAcylation; O-linked N-acetylglucosamine (O-GlcNAc) transferase (OGT); X-linked Intellectual Disability; glycobiology; glycosyltransferase; metabolic disease.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


References
-
- Jackson S. P., and Tjian R. (1988) O-Glycosylation of eukaryotic transcription factors: implications for mechanisms of transcriptional regulation. Cell 55, 125–133 - PubMed
-
- Hanover J. A., Cohen C. K., Willingham M. C., and Park M. K. (1987) O-Linked N-acetylglucosamine is attached to proteins of the nuclear pore: evidence for cytoplasmic and nucleoplasmic glycoproteins. J. Biol. Chem. 262, 9887–9894 - PubMed
-
- Yang W. H., Kim J. E., Nam H. W., Ju J. W., Kim H. S., Kim Y. S., and Cho J. W. (2006) Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat. Cell Biol. 8, 1074–1083 - PubMed
-
- Sayat R., Leber B., Grubac V., Wiltshire L., and Persad S. (2008) O-GlcNAc-glycosylation of β-catenin regulates its nuclear localization and transcriptional activity. Exp. Cell Res. 314, 2774–2787 - PubMed
-
- Torres C. R., and Hart G. W. (1984) Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes: evidence for O-linked GlcNAc. J. Biol. Chem. 259, 3308–3317 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

