Establishment of a vernalization requirement in Brachypodium distachyon requires REPRESSOR OF VERNALIZATION1
- PMID: 28584114
- PMCID: PMC5488934
- DOI: 10.1073/pnas.1700536114
Establishment of a vernalization requirement in Brachypodium distachyon requires REPRESSOR OF VERNALIZATION1
Abstract
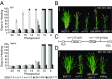
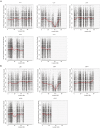
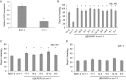
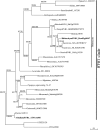
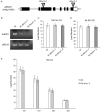
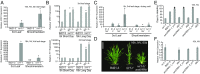
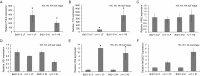
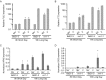
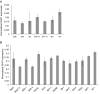
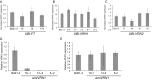
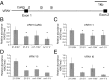
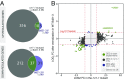
A requirement for vernalization, the process by which prolonged cold exposure provides competence to flower, is an important adaptation to temperate climates that ensures flowering does not occur before the onset of winter. In temperate grasses, vernalization results in the up-regulation of VERNALIZATION1 (VRN1) to establish competence to flower; however, little is known about the mechanism underlying repression of VRN1 in the fall season, which is necessary to establish a vernalization requirement. Here, we report that a plant-specific gene containing a bromo-adjacent homology and transcriptional elongation factor S-II domain, which we named REPRESSOR OF VERNALIZATION1 (RVR1), represses VRN1 before vernalization in Brachypodium distachyon That RVR1 is upstream of VRN1 is supported by the observations that VRN1 is precociously elevated in an rvr1 mutant, resulting in rapid flowering without cold exposure, and the rapid-flowering rvr1 phenotype is dependent on VRN1 The precocious VRN1 expression in rvr1 is associated with reduced levels of the repressive chromatin modification H3K27me3 at VRN1, which is similar to the reduced VRN1 H3K27me3 in vernalized plants. Furthermore, the transcriptome of vernalized wild-type plants overlaps with that of nonvernalized rvr1 plants, indicating loss of rvr1 is similar to the vernalized state at a molecular level. However, loss of rvr1 results in more differentially expressed genes than does vernalization, indicating that RVR1 may be involved in processes other than vernalization despite a lack of any obvious pleiotropy in the rvr1 mutant. This study provides an example of a role for this class of plant-specific genes.
Keywords: Brachypodium; RVR1; VRN1; flowering; vernalization.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Interaction of photoperiod and vernalization determines flowering time of Brachypodium distachyon.Plant Physiol. 2014 Feb;164(2):694-709. doi: 10.1104/pp.113.232678. Epub 2013 Dec 19. Plant Physiol. 2014. PMID: 24357601 Free PMC article.
-
An ortholog of CURLY LEAF/ENHANCER OF ZESTE like-1 is required for proper flowering in Brachypodium distachyon.Plant J. 2018 Mar;93(5):871-882. doi: 10.1111/tpj.13815. Epub 2018 Feb 7. Plant J. 2018. PMID: 29314414
-
A Flowering Locus C Homolog Is a Vernalization-Regulated Repressor in Brachypodium and Is Cold Regulated in Wheat.Plant Physiol. 2017 Feb;173(2):1301-1315. doi: 10.1104/pp.16.01161. Epub 2016 Dec 29. Plant Physiol. 2017. PMID: 28034954 Free PMC article.
-
The molecular basis of vernalization in different plant groups.Cold Spring Harb Symp Quant Biol. 2012;77:105-15. doi: 10.1101/sqb.2013.77.014449. Epub 2013 Apr 25. Cold Spring Harb Symp Quant Biol. 2012. PMID: 23619014 Review.
-
Vernalization and epigenetics: how plants remember winter.Curr Opin Plant Biol. 2004 Feb;7(1):4-10. doi: 10.1016/j.pbi.2003.11.010. Curr Opin Plant Biol. 2004. PMID: 14732435 Review.
Cited by
-
Contemplation on wheat vernalization.Front Plant Sci. 2023 Jan 6;13:1093792. doi: 10.3389/fpls.2022.1093792. eCollection 2022. Front Plant Sci. 2023. PMID: 36684728 Free PMC article. Review.
-
Transcription factor VRT2 reinitiates vernalization when interrupted by warm temperatures in a temperate grass model.Plant Physiol. 2024 Dec 2;196(4):2614-2624. doi: 10.1093/plphys/kiae498. Plant Physiol. 2024. PMID: 39316702 Free PMC article.
-
A florigen paralog is required for short-day vernalization in a pooid grass.Elife. 2019 Jan 8;8:e42153. doi: 10.7554/eLife.42153. Elife. 2019. PMID: 30618375 Free PMC article.
-
EARLY FLOWERING 3 interactions with PHYTOCHROME B and PHOTOPERIOD1 are critical for the photoperiodic regulation of wheat heading time.PLoS Genet. 2023 May 10;19(5):e1010655. doi: 10.1371/journal.pgen.1010655. eCollection 2023 May. PLoS Genet. 2023. PMID: 37163495 Free PMC article.
-
PHYTOCHROME C regulation of photoperiodic flowering via PHOTOPERIOD1 is mediated by EARLY FLOWERING 3 in Brachypodium distachyon.PLoS Genet. 2023 May 10;19(5):e1010706. doi: 10.1371/journal.pgen.1010706. eCollection 2023 May. PLoS Genet. 2023. PMID: 37163541 Free PMC article.
References
-
- Chouard P. Vernalization and its relations to dormancy. Annu Rev Plant Physiol. 1960;11:191–238.
-
- Amasino R. Seasonal and developmental timing of flowering. Plant J. 2010;61:1001–1013. - PubMed
-
- Distelfeld A, Li C, Dubcovsky J. Regulation of flowering in temperate cereals. Curr Opin Plant Biol. 2009;12:178–184. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

