Bcl11b and combinatorial resolution of cell fate in the T-cell gene regulatory network
- PMID: 28584128
- PMCID: PMC5468679
- DOI: 10.1073/pnas.1610617114
Bcl11b and combinatorial resolution of cell fate in the T-cell gene regulatory network
Abstract
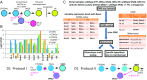
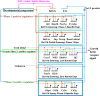
T-cell development from hematopoietic progenitors depends on multiple transcription factors, mobilized and modulated by intrathymic Notch signaling. Key aspects of T-cell specification network architecture have been illuminated through recent reports defining roles of transcription factors PU.1, GATA-3, and E2A, their interactions with Notch signaling, and roles of Runx1, TCF-1, and Hes1, providing bases for a comprehensively updated model of the T-cell specification gene regulatory network presented herein. However, the role of lineage commitment factor Bcl11b has been unclear. We use self-organizing maps on 63 RNA-seq datasets from normal and perturbed T-cell development to identify functional targets of Bcl11b during commitment and relate them to other regulomes. We show that both activation and repression target genes can be bound by Bcl11b in vivo, and that Bcl11b effects overlap with E2A-dependent effects. The newly clarified role of Bcl11b distinguishes discrete components of commitment, resolving how innate lymphoid, myeloid, and dendritic, and B-cell fate alternatives are excluded by different mechanisms.
Keywords: Bcl11b; E2A; Notch-delta signaling; PU.1; commitment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Ikawa T, et al. An essential developmental checkpoint for production of the T cell lineage. Science. 2010;329(5987):93–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

