Impact of enzymatic digestion on bacterial community composition in CF airway samples
- PMID: 28584706
- PMCID: PMC5452939
- DOI: 10.7717/peerj.3362
Impact of enzymatic digestion on bacterial community composition in CF airway samples
Abstract
Background: Previous studies have demonstrated the importance of DNA extraction methods for molecular detection of Staphylococcus, an important bacterial group in cystic fibrosis (CF). We sought to evaluate the effect of enzymatic digestion (EnzD) prior to DNA extraction on bacterial communities identified in sputum and oropharyngeal swab (OP) samples from patients with CF.
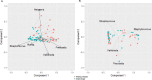
Methods: DNA from 81 samples (39 sputum and 42 OP) collected from 63 patients with CF was extracted in duplicate with and without EnzD. Bacterial communities were determined by rRNA gene sequencing, and measures of alpha and beta diversity were calculated. Principal Coordinate Analysis (PCoA) was used to assess differences at the community level and Wilcoxon Signed Rank tests were used to compare relative abundance (RA) of individual genera for paired samples with and without EnzD.
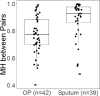
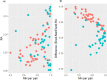
Results: Shannon Diversity Index (alpha-diversity) decreased in sputum and OP samples with the use of EnzD. Larger shifts in community composition were observed for OP samples (beta-diversity, measured by Morisita-Horn), whereas less change in communities was observed for sputum samples. The use of EnzD with OP swabs resulted in significant increase in RA for the genera Gemella (p < 0.01), Streptococcus (p < 0.01), and Rothia (p < 0.01). Staphylococcus (p < 0.01) was the only genus with a significant increase in RA from sputum, whereas the following genera decreased in RA with EnzD: Veillonella (p < 0.01), Granulicatella (p < 0.01), Prevotella (p < 0.01), and Gemella (p = 0.02). In OP samples, higher RA of Gram-positive taxa was associated with larger changes in microbial community composition.
Discussion: We show that the application of EnzD to CF airway samples, particularly OP swabs, results in differences in microbial communities detected by sequencing. Use of EnzD can result in large changes in bacterial community composition, and is particularly useful for detection of Staphylococcus in CF OP samples. The enhanced identification of Staphylococcus aureus is a strong indication to utilize EnzD in studies that use OP swabs to monitor CF airway communities.
Keywords: 16s Sequencing; Beta diversity; DNA extraction; Gram positive; Microbiome; Sputum.
Conflict of interest statement
The authors declare there are no competing interests.
Figures

References
-
- Armstrong DS, Grimwood K, Carlin JB, Carzino R, Olinsky A, Phelan PD. Bronchoalveolar lavage or oropharyngeal cultures to identify lower respiratory pathogens in infants with cystic fibrosis. Pediatric Pulmonology. 1996;21(5):267–275. - PubMed
-
- Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. The Annals of Statistics. 2001;29:1165–1188.
-
- Browder HP, Zygmunt WA, Young JR, Tavormina PA. Lysostaphin: enzymatic mode of action. Biochemical and Biophysical Research Communications. 1965;19:383–389. - PubMed
-
- Burns JL, Emerson J, Stapp JR, Yim DL, Krzewinski J, Louden L, Ramsey BW, Clausen CR. Microbiology of sputum from patients at cystic fibrosis centers in the United States. Clinical Infectious Diseases. 1998;27:158–163. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

