Modeling the cis-regulatory modules of genes expressed in developmental stages of Drosophila melanogaster
- PMID: 28584716
- PMCID: PMC5452948
- DOI: 10.7717/peerj.3389
Modeling the cis-regulatory modules of genes expressed in developmental stages of Drosophila melanogaster
Abstract
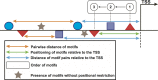
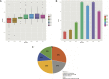
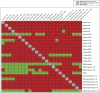
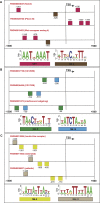
Because transcription is the first step in the regulation of gene expression, understanding how transcription factors bind to their DNA binding motifs has become absolutely necessary. It has been shown that the promoters of genes with similar expression profiles share common structural patterns. This paper presents an extensive study of the regulatory regions of genes expressed in 24 developmental stages of Drosophila melanogaster. It proposes the use of a combination of structural features, such as positioning of individual motifs relative to the transcription start site, orientation, pairwise distance between motifs, and presence of motifs anywhere in the promoter for predicting gene expression from structural features of promoter sequences. RNA-sequencing data was utilized to create and validate the 24 models. When genes with high-scoring promoters were compared to those identified by RNA-seq samples, 19 (79.2%) statistically significant models, a number that exceeds previous studies, were obtained. Each model yielded a set of highly informative features, which were used to search for genes with similar biological functions.
Keywords: Co-expression; Developmental stage; Genetic algorithm; Genome-wide analysis; Promoter architecture; Transcription factor binding sites.
Conflict of interest statement
Kenta Nakai is an Academic Editor for PeerJ.
Figures



Similar articles
-
A set of structural features defines the cis-regulatory modules of antenna-expressed genes in Drosophila melanogaster.PLoS One. 2014 Aug 25;9(8):e104342. doi: 10.1371/journal.pone.0104342. eCollection 2014. PLoS One. 2014. PMID: 25153327 Free PMC article.
-
Using simple rules on presence and positioning of motifs for promoter structure modeling and tissue-specific expression prediction.Genome Inform. 2008;21:188-99. Genome Inform. 2008. PMID: 19425158
-
High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression.Genome Res. 2013 Jan;23(1):169-80. doi: 10.1101/gr.139618.112. Epub 2012 Aug 30. Genome Res. 2013. PMID: 22936248 Free PMC article.
-
TSS seq based core promoter architecture in blood feeding Tsetse fly (Glossina morsitans morsitans) vector of Trypanosomiasis.BMC Genomics. 2015 Sep 22;16(1):722. doi: 10.1186/s12864-015-1921-6. BMC Genomics. 2015. PMID: 26394619 Free PMC article.
-
Structural organization and transcription regulation of nuclear genes encoding the mammalian cytochrome c oxidase complex.Prog Nucleic Acid Res Mol Biol. 1998;61:309-44. doi: 10.1016/s0079-6603(08)60830-2. Prog Nucleic Acid Res Mol Biol. 1998. PMID: 9752724 Review.
Cited by
-
Genetic interactions regulate hypoxia tolerance conferred by activating Notch in excitatory amino acid transporter 1-positive glial cells in Drosophila melanogaster.G3 (Bethesda). 2021 Feb 9;11(2):jkab038. doi: 10.1093/g3journal/jkab038. G3 (Bethesda). 2021. PMID: 33576765 Free PMC article.
References
-
- Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc http://www.bioinformatics.babraham.ac.uk/projects/fastqc
-
- Bajic VB, Choudhary V, Hock CK. Content analysis of the core promoter region of human genes. In Silico Biology. 2003;4:1–15. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

