'Trophic' and 'source' amino acids in trophic estimation: a likely metabolic explanation
- PMID: 28584941
- PMCID: PMC5487837
- DOI: 10.1007/s00442-017-3881-9
'Trophic' and 'source' amino acids in trophic estimation: a likely metabolic explanation
Abstract
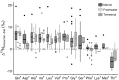
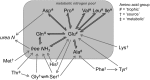
Amino acid nitrogen isotopic analysis is a relatively new method for estimating trophic position. It uses the isotopic difference between an individual's 'trophic' and 'source' amino acids to determine its trophic position. So far, there is no accepted explanation for the mechanism by which the isotopic signals in 'trophic' and 'source' amino acids arise. Yet without a metabolic understanding, the utility of nitrogen isotopic analyses as a method for probing trophic relations, at either bulk tissue or amino acid level, is limited. I draw on isotopic tracer studies of protein metabolism, together with a consideration of amino acid metabolic pathways, to suggest that the 'trophic'/'source' groupings have a fundamental metabolic origin, to do with the cycling of amino-nitrogen between amino acids. 'Trophic' amino acids are those whose amino-nitrogens are interchangeable, part of a metabolic amino-nitrogen pool, and 'source' amino acids are those whose amino-nitrogens are not interchangeable with the metabolic pool. Nitrogen isotopic values of 'trophic' amino acids will reflect an averaged isotopic signal of all such dietary amino acids, offset by the integrated effect of isotopic fractionation from nitrogen cycling, and modulated by metabolic and physiological effects. Isotopic values of 'source' amino acids will be more closely linked to those of equivalent dietary amino acids, but also modulated by metabolism and physiology. The complexity of nitrogen cycling suggests that a single identifiable value for 'trophic discrimination factors' is unlikely to exist. Greater consideration of physiology and metabolism should help in better understanding observed patterns in nitrogen isotopic values.
Keywords: Food web; Isotope; Metabolism; Nitrogen isotopic analysis; Trophic discrimination factor; Trophic enrichment factor.
Figures
Similar articles
-
Beyond bulk δ15N: Combining a suite of stable isotopic measures improves the resolution of the food webs mediating contaminant signals across space, time and communities.Environ Int. 2021 Mar;148:106370. doi: 10.1016/j.envint.2020.106370. Epub 2021 Jan 18. Environ Int. 2021. PMID: 33476789
-
Comparison of δ13C and δ15N of ecologically relevant amino acids among beluga whale tissues.Sci Rep. 2024 May 15;14(1):11146. doi: 10.1038/s41598-024-59307-w. Sci Rep. 2024. PMID: 38750037 Free PMC article.
-
Quantifying capital versus income breeding: New promise with stable isotope measurements of individual amino acids.J Anim Ecol. 2021 Jun;90(6):1408-1418. doi: 10.1111/1365-2656.13402. Epub 2020 Dec 20. J Anim Ecol. 2021. PMID: 33300602
-
Application of amino acids nitrogen stable isotopic analysis in bioaccumulation studies of pollutants: A review.Sci Total Environ. 2023 Jun 25;879:163012. doi: 10.1016/j.scitotenv.2023.163012. Epub 2023 Mar 24. Sci Total Environ. 2023. PMID: 36965734 Review.
-
[Research progress on food sources and food web structure of wetlands based on stable isotopes].Ying Yong Sheng Tai Xue Bao. 2017 Jul 18;28(7):2389-2398. doi: 10.13287/j.1001-9332.201707.027. Ying Yong Sheng Tai Xue Bao. 2017. PMID: 29741074 Review. Chinese.
Cited by
-
Exploring source differences on diet-tissue discrimination factors in the analysis of stable isotope mixing models.Sci Rep. 2020 Sep 25;10(1):15816. doi: 10.1038/s41598-020-73019-x. Sci Rep. 2020. PMID: 32978550 Free PMC article.
-
Amino acid isotope discrimination factors for a carnivore: physiological insights from leopard sharks and their diet.Oecologia. 2018 Dec;188(4):977-989. doi: 10.1007/s00442-018-4276-2. Epub 2018 Oct 22. Oecologia. 2018. PMID: 30349939
-
Investigating long-term trophic stability in North Atlantic cod (Gadus morhua) through nitrogen stable isotope analysis of amino acids.Philos Trans R Soc Lond B Biol Sci. 2025 Jul 10;380(1930):20240028. doi: 10.1098/rstb.2024.0028. Epub 2025 Jul 10. Philos Trans R Soc Lond B Biol Sci. 2025. PMID: 40635432 Free PMC article.
-
Organic Carbon and Nitrogen Isoscapes of Reef Corals and Algal Symbionts: Relative Influences of Environmental Gradients and Heterotrophy.Microorganisms. 2020 Aug 11;8(8):1221. doi: 10.3390/microorganisms8081221. Microorganisms. 2020. PMID: 32796689 Free PMC article.
-
Long-term trends in the foraging ecology and habitat use of an endangered species: an isotopic perspective.Oecologia. 2018 Dec;188(4):1273-1285. doi: 10.1007/s00442-018-4279-z. Epub 2018 Nov 8. Oecologia. 2018. PMID: 30406821
References
-
- Aqvist SEG. Amino acid interrelationships during growth, studied with N15-labeled glycine in regenerating rat liver. Acta Chem Scand. 1951;5:1065–1073. doi: 10.3891/acta.chem.scand.05-1065. - DOI
-
- Aqvist SEG. Metabolic interrelationships among amino acids studied with isotopic nitrogen. Acta Chem Scand. 1951;5:1046–1064. doi: 10.3891/acta.chem.scand.05-1046. - DOI
-
- Bertolo RF, Burrin DG. Comparative aspects of tissue glutamine and proline metabolism. J Nutr. 2008;138:2032S–2039S. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

