One-step generation of complete gene knockout mice and monkeys by CRISPR/Cas9-mediated gene editing with multiple sgRNAs
- PMID: 28585534
- PMCID: PMC5518993
- DOI: 10.1038/cr.2017.81
One-step generation of complete gene knockout mice and monkeys by CRISPR/Cas9-mediated gene editing with multiple sgRNAs
Abstract
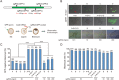
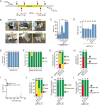
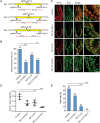
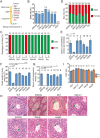
The CRISPR/Cas9 system is an efficient gene-editing method, but the majority of gene-edited animals showed mosaicism, with editing occurring only in a portion of cells. Here we show that single gene or multiple genes can be completely knocked out in mouse and monkey embryos by zygotic injection of Cas9 mRNA and multiple adjacent single-guide RNAs (spaced 10-200 bp apart) that target only a single key exon of each gene. Phenotypic analysis of F0 mice following targeted deletion of eight genes on the Y chromosome individually demonstrated the robustness of this approach in generating knockout mice. Importantly, this approach delivers complete gene knockout at high efficiencies (100% on Arntl and 91% on Prrt2) in monkey embryos. Finally, we could generate a complete Prrt2 knockout monkey in a single step, demonstrating the usefulness of this approach in rapidly establishing gene-edited monkey models.
Figures

References
-
- Niu Y, Shen B, Cui Y, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 2014; 156:836–843. - PubMed
-
- Jennings CG, Landman R, Zhou Y, et al. Opportunities and challenges in modeling human brain disorders in transgenic primates. Nat Neurosci 2016; 19:1123–1130. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

