Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery
- PMID: 28585549
- PMCID: PMC5467206
- DOI: 10.1038/ncomms15790
Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery
Abstract
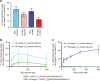
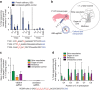
We recently developed base editing, a genome-editing approach that enables the programmable conversion of one base pair into another without double-stranded DNA cleavage, excess stochastic insertions and deletions, or dependence on homology-directed repair. The application of base editing is limited by off-target activity and reliance on intracellular DNA delivery. Here we describe two advances that address these limitations. First, we greatly reduce off-target base editing by installing mutations into our third-generation base editor (BE3) to generate a high-fidelity base editor (HF-BE3). Next, we purify and deliver BE3 and HF-BE3 as ribonucleoprotein (RNP) complexes into mammalian cells, establishing DNA-free base editing. RNP delivery of BE3 confers higher specificity even than plasmid transfection of HF-BE3, while maintaining comparable on-target editing levels. Finally, we apply these advances to deliver BE3 RNPs into both zebrafish embryos and the inner ear of live mice to achieve specific, DNA-free base editing in vivo.
Conflict of interest statement
D.R.L. is a consultant and co-founder of Editas Medicine, a company that seeks to develop genome-editing therapeutics. H.A.R., A.C.K. and D.R.L. have filed patent applications on base editing. The remaining authors declare no competing financial interests.
Figures



References
-
- Doudna J. A. & Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

