Genomics and metagenomics of trimethylamine-utilizing Archaea in the human gut microbiome
- PMID: 28585938
- PMCID: PMC5563959
- DOI: 10.1038/ismej.2017.72
Genomics and metagenomics of trimethylamine-utilizing Archaea in the human gut microbiome
Abstract
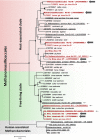
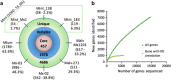
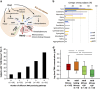
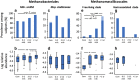
The biological significance of Archaea in the human gut microbiota is largely unclear. We recently reported genomic and biochemical analyses of the Methanomassiliicoccales, a novel order of methanogenic Archaea dwelling in soil and the animal digestive tract. We now show that these Methanomassiliicoccales are present in published microbiome data sets from eight countries. They are represented by five Operational Taxonomic Units present in at least four cohorts and phylogenetically distributed into two clades. Genes for utilizing trimethylamine (TMA), a bacterial precursor to an atherosclerogenic human metabolite, were present in four of the six novel Methanomassiliicoccales genomes assembled from ELDERMET metagenomes. In addition to increased microbiota TMA production capacity in long-term residential care subjects, abundance of TMA-utilizing Methanomassiliicoccales correlated positively with bacterial gene count for TMA production and negatively with fecal TMA concentrations. The two large Methanomassiliicoccales clades have opposite correlations with host health status in the ELDERMET cohort and putative distinct genomic signatures for gut adaptation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Bang C, Schmitz RA. (2015). Archaea associated with human surfaces: not to be underestimated. FEMS Microbiol Rev 39: 631–648. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

