Linking rhizosphere microbiome composition of wild and domesticated Phaseolus vulgaris to genotypic and root phenotypic traits
- PMID: 28585939
- PMCID: PMC5607367
- DOI: 10.1038/ismej.2017.85
Linking rhizosphere microbiome composition of wild and domesticated Phaseolus vulgaris to genotypic and root phenotypic traits
Abstract
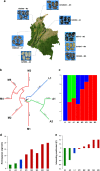
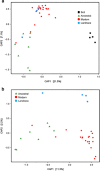
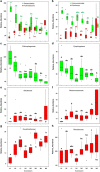
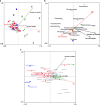
Plant domestication was a pivotal accomplishment in human history, but also led to a reduction in genetic diversity of crop species compared to their wild ancestors. How this reduced genetic diversity affected plant-microbe interactions belowground is largely unknown. Here, we investigated the genetic relatedness, root phenotypic traits and rhizobacterial community composition of modern and wild accessions of common bean (Phaseolus vulgaris) grown in agricultural soil from the highlands of Colombia, one of the centers of common bean diversification. Diversity Array Technology-based genotyping and phenotyping of local common bean accessions showed significant genetic and root architectural differences between wild and modern accessions, with a higher specific root length for the wild accessions. Canonical Correspondence Analysis indicated that the divergence in rhizobacterial community composition between wild and modern bean accessions is associated with differences in specific root length. Along the bean genotypic trajectory, going from wild to modern, we observed a gradual decrease in relative abundance of Bacteroidetes, mainly Chitinophagaceae and Cytophagaceae, and an increase in relative abundance of Actinobacteria and Proteobacteria, in particular Nocardioidaceae and Rhizobiaceae, respectively. Collectively, these results establish a link between common bean domestication, specific root morphological traits and rhizobacterial community assembly.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Akibode S, Maredia M. (2011) Global and regional trends in production, trade and consumption of food legume crops. Report submitted to the CGIAR Standing Panel on Impact Assessment http://www.cgiar.org/our-research/crop-factsheets/beans/).
-
- Berg G, Smalla K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol 68: 1–13. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

