Fast and general tests of genetic interaction for genome-wide association studies
- PMID: 28586362
- PMCID: PMC5478145
- DOI: 10.1371/journal.pcbi.1005556
Fast and general tests of genetic interaction for genome-wide association studies
Abstract
A complex disease has, by definition, multiple genetic causes. In theory, these causes could be identified individually, but their identification will likely benefit from informed use of anticipated interactions between causes. In addition, characterizing and understanding interactions must be considered key to revealing the etiology of any complex disease. Large-scale collaborative efforts are now paving the way for comprehensive studies of interaction. As a consequence, there is a need for methods with a computational efficiency sufficient for modern data sets as well as for improvements of statistical accuracy and power. Another issue is that, currently, the relation between different methods for interaction inference is in many cases not transparent, complicating the comparison and interpretation of results between different interaction studies. In this paper we present computationally efficient tests of interaction for the complete family of generalized linear models (GLMs). The tests can be applied for inference of single or multiple interaction parameters, but we show, by simulation, that jointly testing the full set of interaction parameters yields superior power and control of false positive rate. Based on these tests we also describe how to combine results from multiple independent studies of interaction in a meta-analysis. We investigate the impact of several assumptions commonly made when modeling interactions. We also show that, across the important class of models with a full set of interaction parameters, jointly testing the interaction parameters yields identical results. Further, we apply our method to genetic data for cardiovascular disease. This allowed us to identify a putative interaction involved in Lp(a) plasma levels between two 'tag' variants in the LPA locus (p = 2.42 ⋅ 10-09) as well as replicate the interaction (p = 6.97 ⋅ 10-07). Finally, our meta-analysis method is used in a small (N = 16,181) study of interactions in myocardial infarction.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

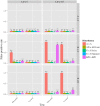

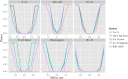

References
-
- Peacock RE, Temple A, Gudnason V, Rosseneu M, Humphries SE. Variation at the lipoprotein lipase and apolipoprotein AI-CIII gene loci are associated with fasting lipid and lipoprotein traits in a population sample from Iceland: interaction between genotype, gender, and smoking status. Genet Epidemiol. 1997;14(3):265–282. 10.1002/(SICI)1098-2272(1997)14:3%3C265::AID-GEPI5%3E3.0.CO;2-4 - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

