Determination of tRNA aminoacylation levels by high-throughput sequencing
- PMID: 28586482
- PMCID: PMC5737633
- DOI: 10.1093/nar/gkx514
Determination of tRNA aminoacylation levels by high-throughput sequencing
Abstract
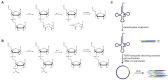
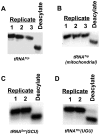
Transfer RNA (tRNA) decodes mRNA codons when aminoacylated (charged) with an amino acid at its 3' end. Charged tRNAs turn over rapidly in cells, and variations in charged tRNA fractions are known to be a useful parameter in cellular responses to stress. tRNA charging fractions can be measured for individual tRNA species using acid denaturing gels, or comparatively at the genome level using microarrays. These hybridization-based approaches cannot be used for high resolution analysis of mammalian tRNAs due to their large sequence diversity. Here we develop a high-throughput sequencing method that enables accurate determination of charged tRNA fractions at single-base resolution (Charged DM-tRNA-seq). Our method takes advantage of the recently developed DM-tRNA-seq method, but includes additional chemical steps that specifically remove the 3'A residue in uncharged tRNA. Charging fraction is obtained by counting the fraction of A-ending reads versus A+C-ending reads for each tRNA species in the same sequencing reaction. In HEK293T cells, most cytosolic tRNAs are charged at >80% levels, whereas tRNASer and tRNAThr are charged at lower levels. These low charging levels were validated using acid denaturing gels. Our method should be widely applicable for investigations of tRNA charging as a parameter in biological regulation.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Elf J., Nilsson D., Tenson T., Ehrenberg M.. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science. 2003; 300:1718–1722. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

