Genome-wide Pleiotropy Between Parkinson Disease and Autoimmune Diseases
- PMID: 28586827
- PMCID: PMC5710535
- DOI: 10.1001/jamaneurol.2017.0469
Genome-wide Pleiotropy Between Parkinson Disease and Autoimmune Diseases
Abstract
Importance: Recent genome-wide association studies (GWAS) and pathway analyses supported long-standing observations of an association between immune-mediated diseases and Parkinson disease (PD). The post-GWAS era provides an opportunity for cross-phenotype analyses between different complex phenotypes.
Objectives: To test the hypothesis that there are common genetic risk variants conveying risk of both PD and autoimmune diseases (ie, pleiotropy) and to identify new shared genetic variants and their pathways by applying a novel statistical framework in a genome-wide approach.
Design, setting, and participants: Using the conjunction false discovery rate method, this study analyzed GWAS data from a selection of archetypal autoimmune diseases among 138 511 individuals of European ancestry and systemically investigated pleiotropy between PD and type 1 diabetes, Crohn disease, ulcerative colitis, rheumatoid arthritis, celiac disease, psoriasis, and multiple sclerosis. NeuroX data (6927 PD cases and 6108 controls) were used for replication. The study investigated the biological correlation between the top loci through protein-protein interaction and changes in the gene expression and methylation levels. The dates of the analysis were June 10, 2015, to March 4, 2017.
Main outcomes and measures: The primary outcome was a list of novel loci and their pathways involved in PD and autoimmune diseases.
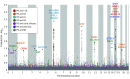
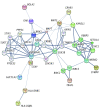
Results: Genome-wide conjunctional analysis identified 17 novel loci at false discovery rate less than 0.05 with overlap between PD and autoimmune diseases, including known PD loci adjacent to GAK, HLA-DRB5, LRRK2, and MAPT for rheumatoid arthritis, ulcerative colitis and Crohn disease. Replication confirmed the involvement of HLA, LRRK2, MAPT, TRIM10, and SETD1A in PD. Among the novel genes discovered, WNT3, KANSL1, CRHR1, BOLA2, and GUCY1A3 are within a protein-protein interaction network with known PD genes. A subset of novel loci was significantly associated with changes in methylation or expression levels of adjacent genes.
Conclusions and relevance: The study findings provide novel mechanistic insights into PD and autoimmune diseases and identify a common genetic pathway between these phenotypes. The results may have implications for future therapeutic trials involving anti-inflammatory agents.
Conflict of interest statement
Figures

Comment in
-
Parkinson Disease and Autoimmune Disorders-What Can We Learn From Genome-wide Pleiotropy?JAMA Neurol. 2017 Jul 1;74(7):769-770. doi: 10.1001/jamaneurol.2017.0843. JAMA Neurol. 2017. PMID: 28586798 No abstract available.
References
-
- Keller MF, Saad M, Bras J, et al. ; International Parkinson’s Disease Genomics Consortium (IPDGC); Wellcome Trust Case Control Consortium 2 (WTCCC2) . Using genome-wide complex trait analysis to quantify “missing heritability” in Parkinson’s disease. Hum Mol Genet. 2012;21(22):4996-5009. - PMC - PubMed
-
- Nalls MA, Pankratz N, Lill CM, et al. ; International Parkinson’s Disease Genomics Consortium (IPDGC); Parkinson’s Study Group (PSG) Parkinson’s Research: The Organized Genetics Initiative (PROGENI); 23andMe; GenePD; NeuroGenetics Research Consortium (NGRC); Hussman Institute of Human Genomics (HIHG); Ashkenazi Jewish Dataset Investigator; Cohorts for Health and Aging Research in Genetic Epidemiology (CHARGE); North American Brain Expression Consortium (NABEC); United Kingdom Brain Expression Consortium (UKBEC); Greek Parkinson’s Disease Consortium; Alzheimer Genetic Analysis Group . Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet. 2014;46(9):989-993. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

