Variability and Global Distribution of Subgenotypes of Bovine Viral Diarrhea Virus
- PMID: 28587150
- PMCID: PMC5490805
- DOI: 10.3390/v9060128
Variability and Global Distribution of Subgenotypes of Bovine Viral Diarrhea Virus
Abstract
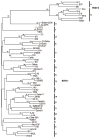
Bovine viral diarrhea virus (BVDV) is a globally-distributed agent responsible for numerous clinical syndromes that lead to major economic losses. Two species, BVDV-1 and BVDV-2, discriminated on the basis of genetic and antigenic differences, are classified in the genus Pestivirus within the Flaviviridae family and distributed on all of the continents. BVDV-1 can be segregated into at least twenty-one subgenotypes (1a-1u), while four subgenotypes have been described for BVDV-2 (2a-2d). With respect to published sequences, the number of virus isolates described for BVDV-1 (88.2%) is considerably higher than for BVDV-2 (11.8%). The most frequently-reported BVDV-1 subgenotype are 1b, followed by 1a and 1c. The highest number of various BVDV subgenotypes has been documented in European countries, indicating greater genetic diversity of the virus on this continent. Current segregation of BVDV field isolates and the designation of subgenotypes are not harmonized. While the species BVDV-1 and BVDV-2 can be clearly differentiated independently from the portion of the genome being compared, analysis of different genomic regions can result in inconsistent assignment of some BVDV isolates to defined subgenotypes. To avoid non-conformities the authors recommend the development of a harmonized system for subdivision of BVDV isolates into defined subgenotypes.
Keywords: bovine viral diarrhea virus; epidemiology; genetic diversity; global distribution; subgenotyping.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Becher P., Avalos-Ramirez R., Orlich M., Cedillo-Rosales S., König M., Schweizer M., Stalder H., Schirrmeier H., Thiel H.J. Genetic and antigenic characterization of novel pestivirus genotypes, implications for classification. Virology. 2003;311:96–104. doi: 10.1016/S0042-6822(03)00192-2. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

