Proteases of Dermatophagoides pteronyssinus
- PMID: 28587273
- PMCID: PMC5486027
- DOI: 10.3390/ijms18061204
Proteases of Dermatophagoides pteronyssinus
Abstract
Since the discovery that Der p 1 is a cysteine protease, the role of proteolytic activity in allergic sensitization has been explored. There are many allergens with proteolytic activity; however, exposure from dust mites is not limited to allergens. In this paper, genomic, transcriptomic and proteomic data on Dermatophagoides pteronyssinus (DP) was mined for information regarding the complete degradome of this house dust mite. D. pteronyssinus has more proteases than the closely related Acari, Dermatophagoides farinae (DF) and Sarcoptes scabiei (SS). The group of proteases in D. pteronyssinus is found to be more highly transcribed than the norm for this species. The distribution of protease types is dominated by the cysteine proteases like Der p 1 that account for about half of protease transcription by abundance, and Der p 1 in particular accounts for 22% of the total protease transcripts. In an analysis of protease stability, the group of allergens (Der p 1, Der p 3, Der p 6, and Der p 9) is found to be more stable than the mean. It is also statistically demonstrated that the protease allergens are simultaneously more highly expressed and more stable than the group of D. pteronyssinus proteases being examined, consistent with common assumptions about allergens in general. There are several significant non-allergen outliers from the normal group of proteases with high expression and high stability that should be examined for IgE binding. This paper compiles the first holistic picture of the D. pteronyssinus degradome to which humans may be exposed.
Keywords: allergen source-derived proteases; allergens; allergic sensitization; exposure; proteases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


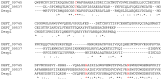

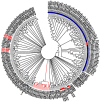
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

